Analysis of the C-terminal membrane anchor domains of hepatitis C virus glycoproteins E1 and E2: toward a topological model
- PMID: 11799189
- PMCID: PMC135876
- DOI: 10.1128/jvi.76.4.1944-1958.2002
Analysis of the C-terminal membrane anchor domains of hepatitis C virus glycoproteins E1 and E2: toward a topological model
Abstract
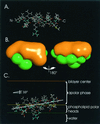
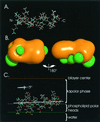
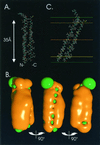
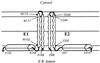
The hepatitis C virus (HCV) glycoproteins E1 and E2 should be anchored in the viral membrane by their C-terminal domains. During synthesis, they are translocated to the endoplasmic reticulum (ER) lumen where they remain. The 31 C-terminal residues of the E1 protein and the 29 C-terminal residues of the E2 protein are implicated in the ER retention. Moreover, the E1 and E2 C termini are implicated in E1-E2 heterodimerization. We studied the E1 and E2 C-terminal sequences of 25 HCV strains in silico using molecular modeling techniques. We conclude that both C-terminal domains should adopt a similar and peculiar configuration: one amphipathic alpha-helix followed by a pair of transmembrane beta-strands. Several three-dimensional (3-D) models were generated. After energy minimization, their ability to interact with membranes was studied using the molecular hydrophobicity potentials calculation and the IMPALA procedure. The latter simulates interactions with a membrane by a Monte Carlo minimization of energy. These methods suggest that the beta-hairpins could anchor the glycoproteins in the ER membrane at least transiently. Anchoring could be stabilized by the adsorption of the nearby amphipathic alpha-helices at the membrane surface. The 3-D models correlate with experimental results which indicate that the E1-E2 transmembrane domains are involved in the heterodimerization and have ER retention properties.
Figures

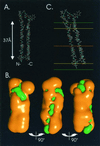
Similar articles
-
A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2.J Virol. 1998 Mar;72(3):2183-91. doi: 10.1128/JVI.72.3.2183-2191.1998. J Virol. 1998. PMID: 9499075 Free PMC article.
-
Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins.J Virol. 2000 Apr;74(8):3623-33. doi: 10.1128/jvi.74.8.3623-3633.2000. J Virol. 2000. PMID: 10729138 Free PMC article.
-
Topological changes in the transmembrane domains of hepatitis C virus envelope glycoproteins.EMBO J. 2002 Jun 17;21(12):2893-902. doi: 10.1093/emboj/cdf295. EMBO J. 2002. PMID: 12065403 Free PMC article.
-
Topology of hepatitis C virus envelope glycoproteins.Rev Med Virol. 2003 Jul-Aug;13(4):233-41. doi: 10.1002/rmv.391. Rev Med Virol. 2003. PMID: 12820185 Review.
-
[Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins].Ann Biol Clin (Paris). 2007 May-Jun;65(3):237-46. Ann Biol Clin (Paris). 2007. PMID: 17502294 Review. French.
Cited by
-
GB virus type C interactions with HIV: the role of envelope glycoproteins.J Viral Hepat. 2009 Nov;16(11):757-68. doi: 10.1111/j.1365-2893.2009.01194.x. Epub 2009 Sep 15. J Viral Hepat. 2009. PMID: 19758271 Free PMC article. Review.
-
Preferential association of Hepatitis C virus with apolipoprotein B48-containing lipoproteins.J Gen Virol. 2006 Oct;87(Pt 10):2983-2991. doi: 10.1099/vir.0.82033-0. J Gen Virol. 2006. PMID: 16963757 Free PMC article.
-
Expression of unmodified hepatitis C virus envelope glycoprotein-coding sequences leads to cryptic intron excision and cell surface expression of E1/E2 heterodimers comprising full-length and partially deleted E1.J Virol. 2003 Dec;77(24):13418-24. doi: 10.1128/jvi.77.24.13418-13424.2003. J Virol. 2003. PMID: 14645599 Free PMC article.
-
A computational approach identifies two regions of Hepatitis C Virus E1 protein as interacting domains involved in viral fusion process.BMC Struct Biol. 2009 Jul 29;9:48. doi: 10.1186/1472-6807-9-48. BMC Struct Biol. 2009. PMID: 19640267 Free PMC article.
-
Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles.J Virol. 2002 Aug;76(15):7672-82. doi: 10.1128/jvi.76.15.7672-7682.2002. J Virol. 2002. PMID: 12097581 Free PMC article.
References
-
- Aggeli, A., N. Boden, Y. L. Cheng, J. B. Findlay, P. F. Knowles, P. Kovatchev, and P. J. Turnbull. 1996. Peptides modeled on the transmembrane region of the slow voltage-gated IsK potassium channel: structural characterization of peptide assemblies in the beta-strand conformation. Biochemistry 35:16213-16221. - PubMed
-
- Alter, M. J., S. C. Hadler, F. N. Judson, A. Mares, W. J. Alexander, P. Y. Hu, J. K. Miller, L. A. Moyer, H. A. Fields, and D. W. Bradley. 1990. Risk factors for acute non-A, non-B hepatitis in the United States and association with hepatitis C virus infection. JAMA 264:2231-2235. - PubMed
-
- Arunkumar, A. I., T. K. Kumar, G. Jayaraman, D. Samuel, and C. Yu. 1996. Induction of helical conformation in all beta-sheet proteins by trifluoroethanol. J. Biomol. Struct. Dyn. 14:381-385. - PubMed
-
- Brasseur, R. 1990. TAMMO: theoretical analysis of membrane molecular organization, p. 203-219. In R. Brasseur (ed.), Molecular description of biological membrane components by computer-aided conformational analysis. CRC Press, Boca Raton, Fla.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

