A sensitive and rapid assay for homologous recombination in mosquito cells: impact of vector topology and implications for gene targeting
- PMID: 11801182
- PMCID: PMC64643
- DOI: 10.1186/1471-2156-2-21
A sensitive and rapid assay for homologous recombination in mosquito cells: impact of vector topology and implications for gene targeting
Abstract
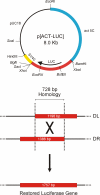
Background: Recent progress in insect transgenesis has been dramatic but existing transposon-based approaches are constrained by position effects and potential instability. Gene targeting would bring a number of benefits, however progress requires a better understanding of the mechanisms involved. Much can be learned in vitro since extrachromosomal recombination occurs at high frequency, facilitating the study of multiple events and the impact of structural changes among the recombining molecules. We have investigated homologous recombination in mosquito cells through restoration of luciferase activity from deleted substrates. The implications of this work for the construction of insect gene targeting vectors are discussed.
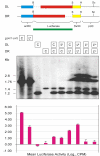
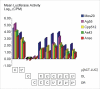
Results: We show that linear targeting vectors are significantly more efficient than circular ones and that recombination is stimulated by introducing double-strand breaks into, or near, the region of homology. Single-strand annealing represents a very efficient pathway but may not be feasible for targeting unbroken chromosomes. Using circular plasmids to mimic chromosomal targets, one-sided invasion appears to be the predominant pathway for homologous recombination. Non-homologous end joining reactions also occur and may be utilised in gene targeting if double-strand breaks are first introduced into the target site.
Conclusions: We describe a rapid, sensitive assay for extrachromosomal homologous recombination in mosquito cells. Variations in substrate topology suggest that single-strand annealing and one-sided invasion represent the predominant pathways, although non-homologous end joining reactions also occur. One-sided invasion of circular chromosomal mimics by linear vectors might therefore be used in vitro to investigate the design and efficiency of gene targeting strategies.
Figures
References
-
- Capecchi MR. Altering the genome by homologous recombination. Science. 1989;16:1288–1292. - PubMed
-
- Morrow B, Kucherlapati R. Gene targeting in mammalian cells by homologous recombination. Curr Opin Biotechnol. 1993;4:577–582. - PubMed
-
- Eggleston P, Zhao Y. Targeted transformation of the insect genome. Insect transgenesis: methods and applications (Edited by Handler AM, James AA) Boca Raton, Florida, CRC Press. 2000. pp. 29–52.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

