Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses
- PMID: 11801185
- PMCID: PMC64570
- DOI: 10.1186/1471-2180-1-34
Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses
Abstract
Background: Post-transcriptional gene silencing (PTGS) by short interfering RNA has opened up new directions in the phenotypic mutation of cellular genes. However, its efficacy on non-nuclear genes and its effect on the interferon pathway remain unexplored. Since directed mutation of RNA genomes is not possible through conventional mutagenesis, we have tested sequence-specific 21-nucleotide long double-stranded RNAs (dsRNAs) for their ability to silence cytoplasmic RNA genomes.
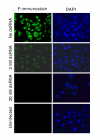
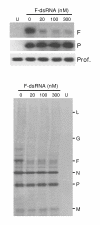
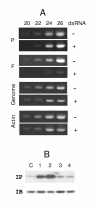
Results: Short dsRNAs were generated against specific mRNAs of respiratory syncytial virus, a nonsegmented negative-stranded RNA virus with a cytoplasmic life cycle. At nanomolar concentrations, the dsRNAs specifically abrogated expression of the corresponding viral proteins, and produced the expected mutant phenotype ex vivo. The dsRNAs did not induce an interferon response, and did not inhibit cellular gene expression. The ablation of the viral proteins correlated with the loss of the specific mRNAs. In contrast, viral genomic and antigenomic RNA, which are encapsidated, were not directly affected.
Conclusions: Synthetic inhibitory dsRNAs are effective in specific silencing of RNA genomes that are exclusively cytoplasmic and transcribed by RNA-dependent RNA polymerases. RNA-directed RNA gene silencing does not require cloning, expression, and mutagenesis of viral cDNA, and thus, will allow the generation of phenotypic null mutants of specific RNA viral genes under normal infection conditions and at any point in the infection cycle. This will, for the first time, permit functional genomic studies, attenuated infections, reverse genetic analysis, and studies of host-virus signaling pathways using a wild type RNA virus, unencumbered by any superinfecting virus.
Figures


References
-
- Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nat Rev Genet. 2001;2:110–119. - PubMed
-
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. - PubMed
-
- Banerjee AK, Barik S, De BP. Gene expression of nonsegmented negative-strand RNA viruses. Pharmacol Ther. 1991;51:47–70. - PubMed
-
- Cogoni C, Macino G. Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev. 2000;10:638–643. - PubMed
-
- Geiss G, Jin G, Guo J, Bumgarner R, Katze MG, Sen GC. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J Biol Chem. 2001;276:30178–30182. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

