Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes
- PMID: 11805320
- PMCID: PMC117357
- DOI: 10.1073/pnas.022531599
Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes
Abstract
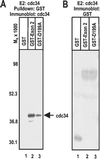
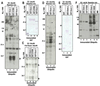
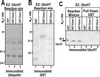
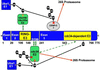
Infected cell protein 0 (ICP0) of herpes simplex virus 1, a multifunctional ring finger protein, enhances the expression of genes introduced into cells by infection or transfection, interacts with numerous cellular and viral proteins, and is associated with the degradation of several cellular proteins. Sequences encoded by exon 2 of ICP0 (residues 20-241) bind the UbcH3 (cdc34) ubiquitin-conjugating enzyme, and its carboxy terminus expresses a ubiquitin ligase activity demonstrable by polyubiquitylation of cdc34 in vitro. We report that: (i) The physical interaction of cdc34 and ICP0 leads to its degradation. Thus, substitution of ICP0 aspartate 199 with alanine attenuates the degradation of cdc34 and its binding to the ICP0 ring finger domain. (ii) Substitution of residue 620 reported to abolish the interaction with a ubiquitin-specific protease has no effect on the function of ubiquitin ligase. (iii) ICP0 contains an additional distinct E3 ligase activity specific for the UbcH5a- and UbcH6 E2-conjugating enzymes mapping to the ring finger domain. This is, to our knowledge, the first identification of a viral protein with at least two physically separated E3 ligase activities with different E2 specificities. The results suggest that each activity may target different proteins.
Figures

References
-
- Roizman B, Knipe D M. In: Fields Virology. 4th Ed. Knipe D M, Howley P, Griffin D E, Lamb R A, Martin M A, Roizman B, Straus S E, editors. New York: Lippincott–Williams & Wilkins; 2001. pp. 2399–2459.
-
- Stow N D, Stow E C. J Gen Virol. 1986;67:2571–2585. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

