ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF
- PMID: 11807076
- PMCID: PMC134800
- DOI: 10.1128/jb.184.4.1155-1162.2002
ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF
Abstract
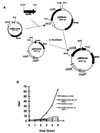
The nosocomial pathogen Enterococcus faecalis has a unique pheromone-inducible conjugative mating system. Conjugative transfer of the E. faecalis plasmid pCF10 is specifically induced by the cCF10 peptide pheromone (LVTLVFV). Genomic sequence information has recently allowed the identification of putative structural genes coding for the various enterococcal pheromones (D. B. Clewell et al., Mol. Microbiol. 35:246-247, 2000). The cCF10 pheromone sequence LVTLVFV was found within an open reading frame designated ccfA, encoding a putative lipoprotein precursor. Several other pheromone sequences were found in similar locations within other predicted lipoproteins. CcfA shows significant sequence relatedness to the Escherichia coli protein YidC, an inner membrane protein translocase, as well as to a large number of homologs identified in gram-positive and in gram-negative bacteria. Analysis of the deduced CcfA amino acid sequence suggested that mature cCF10 peptide could be formed from the proteolytic degradation of its signal peptide. Expression of the cloned ccfA gene with an inducible expression vector dramatically increased cCF10 production by E. faecalis and also resulted in cCF10 production by Lactococcus lactis, a non-pheromone producer. Site-directed mutagenesis of the ccfA sequence encoding the cCF10 peptide confirmed that ccfA was a functional genetic determinant for cCF10.
Figures

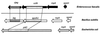


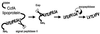
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- An, F. Y., and D. B. Clewell. 1994. Characterization of the determinant (traB) encoding sex pheromone shutdown by the hemolysin/bacteriocin plasmid, pAD1 in Enterococcus faecalis. Plasmid 31:215-221. - PubMed
-
- Anonymous. 2000. TBLASTN, 2.0MP-WashU ed. Washington University, St. Louis, Mo.
-
- Barik, S. 1996. Site-directed mutagenesis in vitro by megaprimer PCR. Methods Mol. Biol. 57:203-215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

