Identification of residues in yeast Spo11p critical for meiotic DNA double-strand break formation
- PMID: 11809802
- PMCID: PMC134631
- DOI: 10.1128/MCB.22.4.1106-1115.2002
Identification of residues in yeast Spo11p critical for meiotic DNA double-strand break formation
Abstract
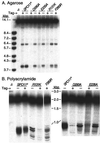
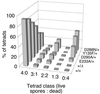
Saccharomyces cerevisiae Spo11 protein (Spo11p) is thought to generate the DNA double-strand breaks (DSBs) that initiate homologous recombination during meiosis. Spo11p is related to a subunit of archaebacterial topoisomerase VI and appears to cleave DNA through a topoisomerase-like transesterase mechanism. In this work, we used the crystal structure of a fragment of topoisomerase VI to model the Spo11p structure and to identify amino acid residues in yeast Spo11p potentially involved in DSB catalysis and/or DNA binding. These residues were mutated to determine which are critical for Spo11p function in vivo. Mutation of Glu-233 or Asp-288, which lie in a conserved structural motif called the Toprim domain, abolished meiotic recombination. These Toprim domain residues have been implicated in binding a metal ion cofactor in topoisomerases and bacterial primases, supporting the idea that DNA cleavage by Spo11p is Mg(2+) dependent. Mutations at an invariant arginine (Arg-131) within a second conserved structural motif known as the 5Y-CAP domain, as well as three other mutations (E235A, F260R, and D290A), caused marked changes in the DSB pattern at a recombination hotspot, suggesting that Spo11p contributes directly to the choice of DNA cleavage site. Finally, certain DSB-defective mutant alleles generated in this study conferred a semidominant negative phenotype but only when Spo11p activity was partially compromised by the presence of an epitope tag. These results are consistent with a multimeric structure for Spo11p in vivo but may also indicate that the amount of Spo11 protein is not a limiting factor for DSB formation in normal cells.
Figures



References
-
- Alani, E., R. Padmore, and N. Kleckner. 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61:419-436. - PubMed
-
- Baudat, F., K. Manova, J. P. Yuen, M. Jasin, and S. Keeney. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6:989-998. - PubMed
-
- Bergerat, A., D. Gadelle, and P. Forterre. 1994. Purification of a DNA topoisomerase II from the hyperthermophilic archaeon Sulfolobus shibatae. A thermostable enzyme with both bacterial and eucaryal features. J. Biol. Chem. 269:27663-27669. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
