A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine
- PMID: 11809803
- PMCID: PMC134642
- DOI: 10.1128/MCB.22.4.1116-1125.2002
A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine
Abstract
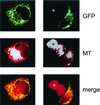
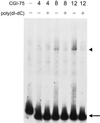
A critical step toward understanding mitochondrial genetics and its impact on human disease is to identify and characterize the full complement of nucleus-encoded factors required for mitochondrial gene expression and mitochondrial DNA (mtDNA) replication. Two factors required for transcription initiation from a human mitochondrial promoter are h-mtRNA polymerase and the DNA binding transcription factor, h-mtTFA. However, based on studies in model systems, the existence of a second human mitochondrial transcription factor has been postulated. Here we report the isolation of a cDNA encoding h-mtTFB, the human homolog of Saccharomyces cerevisiae mitochondrial transcription factor B (sc-mtTFB) and the first metazoan member of this class of transcription factors to which a gene has been assigned. Recombinant h-mtTFB is capable of binding mtDNA in a non-sequence-specific fashion and activates transcription from the human mitochondrial light-strand promoter in the presence of h-mtTFA in vitro. Remarkably, h-mtTFB and its fungal homologs are related in primary sequence to a superfamily of N6 adenine RNA methyltransferases. This observation, coupled with the ability of recombinant h-mtTFB to bind S-adenosylmethionine in vitro, suggests that a structural, and perhaps functional, relationship exists between this class of transcription factors and this family of RNA modification enzymes and that h-mtTFB may perform dual functions during mitochondrial gene expression.
Figures


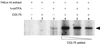


References
-
- Abad-Zapatero, C., P. Zhong, D. E. Bussiere, K. Stewart, and S. W. Muchmore. 1999. rRNA methyltransferases (ErmC" and ErmAM) and antibiotic resistance, p. 199-205. In X. Cheng and R. M. Blumenthal (ed.), S-Adenosylmethionine-dependent methyltransferases: structure and function. World Scientific Publishing, River Edge, N.J.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bogenhagen, D. F. 1996. Interaction of mtTFB and mtRNA polymerase at core promoters for transcription of Xenopus laevis mtDNA. J. Biol. Chem. 271:12036-12041. - PubMed
-
- Bussiere, D. E., S. W. Muchmore, C. G. Dealwis, G. Schluckebier, V. L. Nienaber, R. P. Edalji, K. A. Walter, U. S. Ladror, T. F. Holzman, and C. Abad-Zapatero. 1998. Crystal structure of ErmC", an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry 37:7103-7112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
