The AT-hook protein D1 is essential for Drosophila melanogaster development and is implicated in position-effect variegation
- PMID: 11809812
- PMCID: PMC134649
- DOI: 10.1128/MCB.22.4.1218-1232.2002
The AT-hook protein D1 is essential for Drosophila melanogaster development and is implicated in position-effect variegation
Abstract
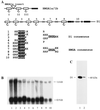
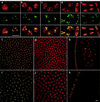
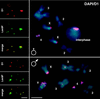
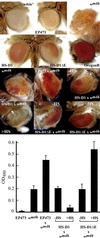
We have analyzed the expression pattern of the D1 gene and the localization of its product, the AT hook-bearing nonhistone chromosomal protein D1, during Drosophila melanogaster development. D1 mRNAs and protein are maternally contributed, and the protein localizes to discrete foci on the chromosomes of early embryos. These foci correspond to 1.672- and 1.688-g/cm(3) AT-rich satellite repeats found in the centromeric heterochromatin of the X and Y chromosomes and on chromosomes 3 and 4. D1 mRNA levels subsequently decrease throughout later development, followed by the accumulation of the D1 protein in adult gonads, where two distributions of D1 can be correlated to different states of gene activity. We show that the EP473 mutation, a P-element insertion upstream of D1 coding sequences, affects the expression of the D1 gene and results in an embryonic homozygous lethal phenotype correlated with the depletion of D1 protein during embryogenesis. Remarkably, decreased levels of D1 mRNA and protein in heterozygous flies lead to the suppression of position-effect variegation (PEV) of the white gene in the white-mottled (w(m4h)) X-chromosome inversion. Our results identify D1 as a DNA-binding protein of known sequence specificity implicated in PEV. D1 is the primary factor that binds the centromeric 1.688-g/cm(3) satellite repeats which are likely involved in white-mottled variegation. We propose that the AT-hook D1 protein nucleates heterochromatin assembly by recruiting specialized transcriptional repressors and/or proteins involved in chromosome condensation.
Figures



References
-
- Alfageme, C. R., G. T. Rudkin, and L. H. Cohen. 1980. Isolation, properties and cellular distribution of D1, a chromosomal protein of Drosophila. Chromosoma 78:1-80. - PubMed
-
- Amirand, C., A. Viari, J. P. Ballini, H. Rezaei, N. Beaujean, D. Jullien, E. Käs, and P. Debey. 1998. Three distinct sub-nuclear populations of HMG-I protein of different properties revealed by colocalization image analysis. J. Cell Sci. 111:3551-3561. - PubMed
-
- Ashley, C. T., C. G. Pendleton, W. W. Jennings, A. Saxena, and C. V. Glover. 1989. Isolation and sequencing of cDNA clones encoding Drosophila chromosomal protein D1. A repeating motif in proteins which recognize at DNA. J. Biol. Chem. 264:8394-8401. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
