A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI
- PMID: 11809876
- PMCID: PMC100309
- DOI: 10.1093/nar/30.3.649
A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI
Abstract
The ATPase ISWI is the catalytic core of several nucleosome remodeling complexes, which are able to alter histone-DNA interactions within nucleosomes such that the sliding of histone octamers on DNA is facilitated. Dynamic nucleosome repositioning may be involved in the assembly of chromatin with regularly spaced nucleosomes and accessible regulatory sequence elements. The mechanism that underlies nucleosome sliding is largely unresolved. We recently discovered that the N-terminal 'tail' of histone H4 is critical for nucleosome remodeling by ISWI. If deleted, nucleosomes are no longer recognized as substrates and do not stimulate the ATPase activity of ISWI. We show here that the H4 tail is part of a more complex recognition epitope which is destroyed by grafting the H4 N-terminus onto other histones. We mapped the H4 tail requirement to a hydrophilic patch consisting of the amino acids R17H18R19 localized at the base of the tail. These residues have been shown earlier to contact nucleosomal DNA, suggesting that ISWI recognizes an 'epitope' consisting of the DNA-bound H4 tail. Consistent with this hypothesis, the ISWI ATPase is stimulated by isolated H4 tail peptides ISWI only in the presence of DNA. Acetylation of the adjacent K12 and K16 residues impairs substrate recognition by ISWI.
Figures


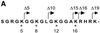
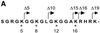



References
-
- Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. - PubMed
-
- Peterson C.L. (2000) ATP-dependent chromatin remodeling: going mobile. FEBS Lett., 476, 68–72. - PubMed
-
- Becker P.B. and Hörz,W. (2002) ATP-dependent nucleosome remodeling. Annu. Rev. Biochem., in press. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

