Conserved economics of transcription termination in eubacteria
- PMID: 11809879
- PMCID: PMC100295
- DOI: 10.1093/nar/30.3.675
Conserved economics of transcription termination in eubacteria
Abstract
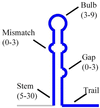
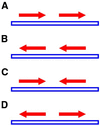
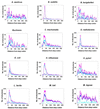
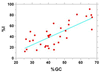
A secondary structure in the nascent RNA followed by a trail of U residues is believed to be necessary and sufficient to terminate transcription. Such structures represent an extremely economical mechanism of transcription termination since they function in the absence of any additional protein factors. We have developed a new algorithm, GeSTer, to identify putative terminators and analysed all available complete bacterial genomes. The algorithm classifies the structures into five classes. We find that potential secondary structure sequences are concentrated downstream of coding regions in most bacterial genomes. Interestingly, many of these structures are not followed by a discernible U-trail. However, irrespective of the nature of the trail sequence, the structures show a similar distribution, indicating that they serve the same purpose. In contrast, such a distribution is absent in archaeal genomes, indicating that they employ a distinct mechanism for transcription termination. The present algorithm represents the fastest and most accurate algorithm for identifying terminators in eubacterial genomes without being restricted by the classical Escherichia coli paradigm.
Figures




References
-
- von Hippel P.H. (1998) An integrated model of the transcription complex in elongation, termination and editing. Science, 281, 660–665. - PubMed
-
- Das A. (1993) Control of transcription termination by RNA-binding proteins. Annu. Rev. Biochem., 62, 893–930. - PubMed
-
- Richardson J.P. and Greenblatt,J. (1996) Control of RNA chain elongation and termination. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd Edn. ASM Press, Washington, DC, pp. 822–848.
-
- Platt T. (1994) Rho and RNA: models for recognition and response. Mol. Microbiol., 11, 983–990. - PubMed
-
- Platt T. (1986) Transcription termination and the regulation of gene expression. Annu. Rev. Biochem., 55, 339–372. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

