Identification, cloning and characterization of a new DNA-binding protein from the hyperthermophilic methanogen Methanopyrus kandleri
- PMID: 11809880
- PMCID: PMC100301
- DOI: 10.1093/nar/30.3.685
Identification, cloning and characterization of a new DNA-binding protein from the hyperthermophilic methanogen Methanopyrus kandleri
Abstract
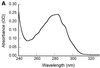
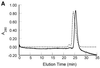
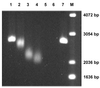
Three novel DNA-binding proteins with apparent molecular masses of 7, 10 and 30 kDa have been isolated from the hyperthermophilic methanogen Methanopyrus kandleri. The proteins were identified using a blot overlay assay that was modified to emulate the high ionic strength intracellular environment of M.kandleri proteins. A 7 kDa protein, named 7kMk, was cloned and expressed in Escherichia coli. As indicated by CD spectroscopy and computer-assisted structure prediction methods, 7kMk is a substantially alpha-helical protein possibly containing a short N-terminal beta-strand. According to analytical gel filtration chromatography and chemical crosslinking, 7kMk exists as a stable dimer, susceptible to further oligomerization. Electron microscopy showed that 7kMk bends DNA and also leads to the formation of loop-like structures of approximately 43.5 +/- 3.5 nm (136 +/- 11 bp for B-form DNA) circumference. A topoisomerase relaxation assay demonstrated that looped DNA is negatively supercoiled under physiologically relevant conditions (high salt and temperature). A BLAST search did not yield 7kMk homologs at the amino acid sequence level, but based on a multiple alignment with ribbon-helix-helix (RHH) transcriptional regulators, fold features and self-association properties of 7kMk we hypothesize that it could be related to RHH proteins.
Figures






References
-
- Sandman K. and Reeve,J.N. (2000) Structure and functional relationships of archaeal and eukaryal histones and nucleosomes. Arch. Microbiol., 173, 165–169. - PubMed
-
- Rowlands T., Baumann,P. and Jackson,S.P. (1994) The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science, 264, 1326–1329. - PubMed
-
- Ouzounis C. and Sander,C. (1992) TFIIB, an evolutionary link between the transcription machineries of archaebacteria and eukaryotes. Cell, 71, 189–190. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

