All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae
- PMID: 11812830
- PMCID: PMC97604
- DOI: 10.1093/nar/29.24.5001
All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae
Abstract
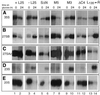
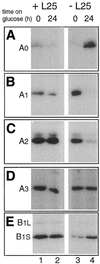
Mutational analysis has shown that the integrity of the region in domain III of 25S rRNA that is involved in binding of ribosomal protein L25 is essential for the production of mature 25S rRNA in the yeast Saccharomyces cerevisiae. However, even structural alterations that do not noticeably affect recognition by L25, as measured by an in vitro assay, strongly reduced 25S rRNA formation by inhibiting the removal of ITS2 from the 27S(B) precursor. In order to analyze the role of L25 in yeast pre-rRNA processing further we studied the effect of genetic depletion of the protein or mutation of each of its three previously identified functional domains, involved in nuclear import (N-terminal), RNA binding (central) and 60S subunit assembly (C-terminal), respectively. Depletion of L25 or mutating its (pre-)rRNA-binding domain blocked conversion of the 27S(B) precursor to 5.8S/25S rRNA, confirming that assembly of L25 is essential for ITS2 processing. However, mutations in either the N- or the C-terminal domain of L25, which only marginally affect its ability to bind to (pre-)rRNA, also resulted in defective ITS2 processing. Furthermore, in all cases there was a notable reduction in the efficiency of processing at the early cleavage sites A0, A1 and A2. We conclude that the assembly of L25 is necessary but not sufficient for removal of ITS2, as well as for fully efficient cleavage at the early sites. Additional elements located in the N- as well as C-terminal domains of L25 are required for both aspects of pre-rRNA processing.
Figures


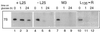


Similar articles
-
Domain III of Saccharomyces cerevisiae 25 S ribosomal RNA: its role in binding of ribosomal protein L25 and 60 S subunit formation.J Mol Biol. 2000 Feb 11;296(1):7-17. doi: 10.1006/jmbi.1999.3432. J Mol Biol. 2000. PMID: 10656814
-
Yeast ribosomal proteins L4, L17, L20, and L25 exhibit different binding characteristics for the yeast 35S precursor rRNA.Biochim Biophys Acta. 1998 Nov 26;1443(1-2):139-48. doi: 10.1016/s0167-4781(98)00202-4. Biochim Biophys Acta. 1998. PMID: 9838082
-
The Role of Ribosomal Proteins eL15 and eL36 in the Early Steps of Yeast 60S Ribosomal Subunit Assembly.J Mol Biol. 2023 Dec 15;435(24):168321. doi: 10.1016/j.jmb.2023.168321. Epub 2023 Oct 20. J Mol Biol. 2023. PMID: 37865285
-
Caught in the act-Visualizing ribonucleases during eukaryotic ribosome assembly.Wiley Interdiscip Rev RNA. 2023 Jul-Aug;14(4):e1766. doi: 10.1002/wrna.1766. Epub 2022 Oct 18. Wiley Interdiscip Rev RNA. 2023. PMID: 36254602 Review.
-
Putting It All Together: The Roles of Ribosomal Proteins in Nucleolar Stages of 60S Ribosomal Assembly in the Yeast Saccharomyces cerevisiae.Biomolecules. 2024 Aug 9;14(8):975. doi: 10.3390/biom14080975. Biomolecules. 2024. PMID: 39199362 Free PMC article. Review.
Cited by
-
rRNA maturation in yeast cells depleted of large ribosomal subunit proteins.PLoS One. 2009 Dec 11;4(12):e8249. doi: 10.1371/journal.pone.0008249. PLoS One. 2009. PMID: 20011513 Free PMC article.
-
Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low-molecular-mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60S ribosomal subunit biogenesis.Mol Cell Biol. 2007 Feb;27(4):1207-21. doi: 10.1128/MCB.01523-06. Epub 2006 Dec 4. Mol Cell Biol. 2007. PMID: 17145778 Free PMC article.
-
Ribosome biogenesis in the yeast Saccharomyces cerevisiae.Genetics. 2013 Nov;195(3):643-81. doi: 10.1534/genetics.113.153197. Genetics. 2013. PMID: 24190922 Free PMC article. Review.
-
Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development.Plant Physiol. 2008 May;147(1):128-42. doi: 10.1104/pp.107.111799. Epub 2008 Mar 5. Plant Physiol. 2008. PMID: 18322146 Free PMC article.
-
Decoding Plant Ribosomal Proteins: Multitasking Players in Cellular Games.Cells. 2025 Mar 21;14(7):473. doi: 10.3390/cells14070473. Cells. 2025. PMID: 40214427 Free PMC article. Review.
References
-
- Tollervey D. (1996) Trans-acting factors in ribosome synthesis. Exp. Cell Res., 229, 226–232. - PubMed
-
- Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 216–311. - PubMed
-
- Bachellerie J.-P., Cavaillé,J. and Qu,L.-H. (2000) Nucleotide modifications of eukaryotic rRNAs: the world of small nucleolar RNAs guides revisited. In Garrett,R.A., Douthwaite,S.A., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. American Society for Microbiology, Washington, DC, pp. 191–204.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

