An architectural role of the Escherichia coli chromatin protein FIS in organising DNA
- PMID: 11812843
- PMCID: PMC97572
- DOI: 10.1093/nar/29.24.5107
An architectural role of the Escherichia coli chromatin protein FIS in organising DNA
Abstract
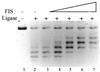
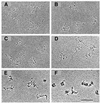
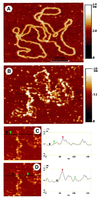
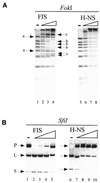
The Escherichia coli chromatin protein FIS modulates the topology of DNA in a growth phase-dependent manner. In this study we have investigated the global effect of FIS binding on DNA architecture in vitro. We show that in supercoiled DNA molecules FIS binds at multiple sites in a non-random fashion and increases DNA branching. This global DNA reshaping effect is independent of the helical phasing of FIS binding sites. We propose, in addition to the previously inferred stabilisation of tightly bent DNA microloops in the upstream regions of certain promoters, that FIS may perform the distinct architectural function of organising branched plectonemes in the E.coli nucleoid.
Figures


References
-
- Sinden R.R., Carlson,J.O. and Pettijohn,D.E. (1980) Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell, 21, 773–783. - PubMed
-
- Boles T.C., White,J.H. and Cozzarelli,N.R. (1990) Structure of plectonemically supercoiled DNA. J. Mol. Biol., 213, 931–951. - PubMed
-
- Worcel A. and Burgi,E. (1972) On the structure of the folded chromosome of Escherichia coli.J. Mol. Biol., 71, 127–147. - PubMed
-
- Staczek P. and Higgins,N.P. (1998) Gyrase and topo IV modulate chromosome domain size in vivo. Mol. Microbiol., 29, 1435–1448. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

