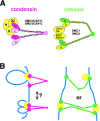
Condensin and cohesin display different arm conformations with characteristic hinge angles
- PMID: 11815634
- PMCID: PMC2173330
- DOI: 10.1083/jcb.200111002
Condensin and cohesin display different arm conformations with characteristic hinge angles
Abstract
Structural maintenance of chromosomes (SMC) proteins play central roles in higher-order chromosome dynamics from bacteria to humans. In eukaryotes, two different SMC protein complexes, condensin and cohesin, regulate chromosome condensation and sister chromatid cohesion, respectively. Each of the complexes consists of a heterodimeric pair of SMC subunits and two or three non-SMC subunits. Previous studies have shown that a bacterial SMC homodimer has a symmetrical structure in which two long coiled-coil arms are connected by a flexible hinge. A catalytic domain with DNA- and ATP-binding activities is located at the distal end of each arm. We report here the visualization of vertebrate condensin and cohesin by electron microscopy. Both complexes display the two-armed structure characteristic of SMC proteins, but their conformations are remarkably different. The hinge of condensin is closed and the coiled-coil arms are placed close together. In contrast, the hinge of cohesin is wide open and the coiled-coils are spread apart from each other. The non-SMC subunits of both condensin and cohesin form a globular complex bound to the catalytic domains of the SMC heterodimers. We propose that the "closed" conformation of condensin and the "open" conformation of cohesin are important structural properties that contribute to their specialized biochemical and physiological functions.
Figures



References
-
- Anderson, D.E., K.M. Trujillo, P. Sung, and H.P. Erickson. 2001. Structure of the Rad50/Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J. Biol. Chem. 276:37027–37033. - PubMed
-
- Cobbe, N., and M.M. Heck. 2000. SMCs in the world of chromosome biology: from prokaryotes to higher eukaryotes. J. Struct. Biol. 129:123–143. - PubMed
-
- Dej, K.J., and T.L. Orr-Weaver. 2000. Separation anxiety at the centromere. Trends Cell Biol. 10:392–399. - PubMed
-
- Fowler, W.E., and H.P. Erickson. 1979. Trinodular structure of fibrinogen. Confirmation by both shadowing and negative stain electron microscopy. J. Mol. Biol. 134:241–249. - PubMed

