The regulatory role for the ERCC3 helicase of general transcription factor TFIIH during promoter escape in transcriptional activation
- PMID: 11818577
- PMCID: PMC122168
- DOI: 10.1073/pnas.251674198
The regulatory role for the ERCC3 helicase of general transcription factor TFIIH during promoter escape in transcriptional activation
Abstract
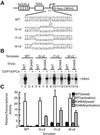
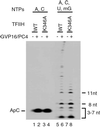
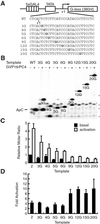
Eukaryotic transcriptional activators have been proposed to function, for the most part, by promoting the assembly of preinitiation complex through the recruitment of the RNA polymerase II transcriptional machinery to the promoter. Previous studies have shown that transcriptional activation is critically dependent on transcription factor IIH (TFIIH), which functions during promoter opening and promoter escape, the steps following preinitiation complex assembly. Here we have analyzed the role of TFIIH in transcriptional activation and show that the excision repair cross-complementing (ERCC) 3 helicase activity of TFIIH plays a regulatory role to stimulate promoter escape in activated transcription. The stimulatory effect of the ERCC3 helicase is observed until approximately 10-nt RNA is synthesized, and the helicase seems to act throughout the entire course of promoter escape. Analyses of the early phase of transcription show that a majority of the initiated complexes abort transcription and fail to escape the promoter; however, the proportion of productive complexes that escape the promoter apparently increases in response to activation. Our results establish that promoter escape is an important regulatory step stimulated by the ERCC3 helicase activity in response to activation and reveal a possible mechanism of transcriptional synergy.
Figures




Similar articles
-
Recruitment of TBP or TFIIB to a promoter proximal position leads to stimulation of RNA polymerase II transcription without activator proteins both in vivo and in vitro.Biochem Biophys Res Commun. 1999 Mar 5;256(1):45-51. doi: 10.1006/bbrc.1999.0280. Biochem Biophys Res Commun. 1999. PMID: 10066420
-
Promoter escape by RNA polymerase II. Formation of an escape-competent transcriptional intermediate is a prerequisite for exit of polymerase from the promoter.J Biol Chem. 1997 Nov 7;272(45):28175-8. doi: 10.1074/jbc.272.45.28175. J Biol Chem. 1997. PMID: 9353262
-
Rad25p, a DNA helicase subunit of yeast transcription factor TFIIH, is required for promoter escape in vivo.Gene. 2000 Mar 7;245(1):109-17. doi: 10.1016/s0378-1119(00)00029-9. Gene. 2000. PMID: 10713451
-
Mechanism of transcription initiation and promoter escape by RNA polymerase II.Curr Opin Genet Dev. 2001 Apr;11(2):209-14. doi: 10.1016/s0959-437x(00)00181-7. Curr Opin Genet Dev. 2001. PMID: 11250146 Review.
-
How eukaryotic transcription activators increase assembly of preinitiation complexes.Cold Spring Harb Symp Quant Biol. 1993;58:199-203. doi: 10.1101/sqb.1993.058.01.024. Cold Spring Harb Symp Quant Biol. 1993. PMID: 7956030 Review. No abstract available.
Cited by
-
Transcriptional coactivator PC4 stimulates promoter escape and facilitates transcriptional synergy by GAL4-VP16.Mol Cell Biol. 2004 Jul;24(14):6525-35. doi: 10.1128/MCB.24.14.6525-6535.2004. Mol Cell Biol. 2004. PMID: 15226451 Free PMC article.
-
Signal transduction pathways controlling Ins2 gene activity and beta cell state transitions.iScience. 2025 Feb 17;28(3):112015. doi: 10.1016/j.isci.2025.112015. eCollection 2025 Mar 21. iScience. 2025. PMID: 40144638 Free PMC article.
-
Defective Hfp-dependent transcriptional repression of dMYC is fundamental to tissue overgrowth in Drosophila XPB models.Nat Commun. 2015 Jun 15;6:7404. doi: 10.1038/ncomms8404. Nat Commun. 2015. PMID: 26074141 Free PMC article.
-
Stimulation of the XPB ATP-dependent helicase by the beta subunit of TFIIE.Nucleic Acids Res. 2005 May 25;33(9):3072-81. doi: 10.1093/nar/gki623. Print 2005. Nucleic Acids Res. 2005. PMID: 15917439 Free PMC article.
-
Extensive transcriptome analysis correlates the plasticity of Entamoeba histolytica pathogenesis to rapid phenotype changes depending on the environment.Sci Rep. 2016 Oct 21;6:35852. doi: 10.1038/srep35852. Sci Rep. 2016. PMID: 27767091 Free PMC article.
References
-
- Triezenberg S J. Curr Opin Genet Dev. 1995;5:190–196. - PubMed
-
- Roeder R G. Trends Biochem Sci. 1996;21:327–335. - PubMed
-
- Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. - PubMed
-
- Kugel J F, Goodrich J A. J Biol Chem. 2000;275:40483–40493. - PubMed
-
- Ptashne M, Gann A. Nature (London) 1997;386:569–577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

