Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex
- PMID: 11823428
- PMCID: PMC125831
- DOI: 10.1093/emboj/21.3.355
Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex
Abstract
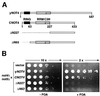
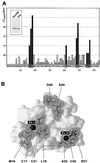
The RING finger protein CNOT4 is a component of the CCR4-NOT complex. This complex is implicated in repression of RNA polymerase II transcription. Here we demonstrate that CNOT4 functions as a ubiquitin-protein ligase (E3). We show that the unique C4C4 RING domain of CNOT4 interacts with a subset of ubiquitin-conjugating enzymes (E2s). Using NMR spectroscopy, we detail the interaction of CNOT4 with UbcH5B and characterize RING residues that are critical for this interaction. CNOT4 acts as a potent E3 ligase in vitro. Mutations that destabilize the E2-E3 interface abolish this activity. Based on these results, we present a model of how E3 ligase function within the CCR4-NOT complex relates to transcriptional regulation.
Figures





References
-
- Albright S.R. and Tjian,R. (2000) TAFs revisited: more data reveal twists and confirm old ideas. Gene, 242, 1–13. - PubMed
-
- Barlow P.N., Luisi,B., Milner,A., Elliott,M. and Everett,R. (1994) Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J. Mol. Biol., 237, 201–211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

