Transvection effects involving DNA methylation during meiosis in the mouse
- PMID: 11823436
- PMCID: PMC125843
- DOI: 10.1093/emboj/21.3.440
Transvection effects involving DNA methylation during meiosis in the mouse
Abstract
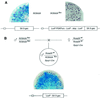
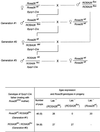
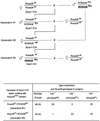
High efficiencies of recombination between LoxP elements were initially recorded when the Cre recombinase was expressed in meiotic spermatocytes. However, it was unexpectedly found that LoxP recombination fell to very low values at the second generation of mice expressing Cre during meiosis. The inability of the LoxP elements to serve as recombination substrates was correlated with cytosine methylation, initially in LoxP and transgene sequences, but later extending for distances of at least several kilobases into chromosomal sequences. It also affected the allelic locus, implying a transfer of structural information between alleles similar to the transvection phenomenon described in Drosophila. Once initiated following Cre-LoxP interaction, neither cis-extension nor transvection of the methylated state required the continuous expression of Cre, as they occurred both in germinal and somatic cells and in the fraction of the offspring that had not inherited the Sycp1-Cre transgene. Therefore, these processes depend on a physiological mechanism of establishment and extension of an epigenetic state, for which they provide an experimental model.
Figures
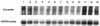

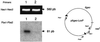





References
-
- Aramayo R. and Metzenberg,R.L. (1996) Meiotic transvection in fungi. Cell, 86, 103–113. - PubMed
-
- Brennan C.A., Van Cleve,M.D. and Gumport,R.I. (1986) The effects of base analogue substitutions on the cleavage by the EcoRI restriction endonuclease of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J. Biol. Chem., 261, 7270–7278. - PubMed
-
- Chomczynski P. and Sacchi,S. (1987) Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 156–159. - PubMed
-
- Colot V. and Rossignol,J.L. (1999) Eukaryotic DNA methylation as an evolutionary device. BioEssays, 21, 402–411. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

