Identification of Mycobacterium spp. by using a commercial 16S ribosomal DNA sequencing kit and additional sequencing libraries
- PMID: 11825949
- PMCID: PMC153382
- DOI: 10.1128/JCM.40.2.400-406.2002
Identification of Mycobacterium spp. by using a commercial 16S ribosomal DNA sequencing kit and additional sequencing libraries
Abstract
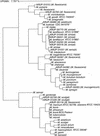
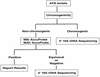
Current methods for identification of Mycobacterium spp. rely upon time-consuming phenotypic tests, mycolic acid analysis, and narrow-spectrum nucleic acid probes. Newer approaches include PCR and sequencing technologies. We evaluated the MicroSeq 500 16S ribosomal DNA (rDNA) bacterial sequencing kit (Applied Biosystems, Foster City, Calif.) for its ability to identify Mycobacterium isolates. The kit is based on PCR and sequencing of the first 500 bp of the bacterial rRNA gene. One hundred nineteen mycobacterial isolates (94 clinical isolates and 25 reference strains) were identified using traditional phenotypic methods and the MicroSeq system in conjunction with separate databases. The sequencing system gave 87% (104 of 119) concordant results when compared with traditional phenotypic methods. An independent laboratory using a separate database analyzed the sequences of the 15 discordant samples and confirmed the results. The use of 16S rDNA sequencing technology for identification of Mycobacterium spp. provides more rapid and more accurate characterization than do phenotypic methods. The MicroSeq 500 system simplifies the sequencing process but, in its present form, requires use of additional databases such as the Ribosomal Differentiation of Medical Microorganisms (RIDOM) to precisely identify subtypes of type strains and species not currently in the MicroSeq library.
Figures
References
-
- American Thoracic Society. 1997. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Am. J. Respir. Crit. Care Med. 156:S1-S25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

