Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load
- PMID: 11825954
- PMCID: PMC153363
- DOI: 10.1128/JCM.40.2.439-445.2002
Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load
Abstract
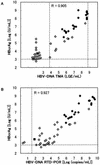
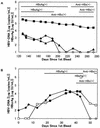
A sensitive enzyme immunoassay (EIA) specific for hepatitis B virus core antigen (HBcAg) and hepatitis B e antigen (HBeAg) was developed. We designated the precore/core gene products as hepatitis B virus (HBV) core-related antigens (HBcrAg). In order to detect HBcrAg even in anti-HBc/e antibody-positive specimens, the specimens were pretreated in detergents. The antibodies are inactivated by this pretreatment and, simultaneously, the antigens are released and the epitopes are exposed. The assay demonstrated 71 to 112% recovery using HBcrAg-positive sera. We observed no interference from the tested anticoagulants or blood components. When the cutoff value was tentatively set at 10(3) U/ml, all healthy control (HBsAg/HBV-DNA negative; n = 108) and anti-HCV antibody-positive (n = 59) sera were identified as negative. The assay showed a detection limit of 4 x 10(2) U/ml using recombinant antigen. Detection limits were compared in four serially diluted HBV high-titer sera. The HBcrAg assay demonstrated higher sensitivity than HBV-DNA transcription-mediated amplification (TMA) or HBeAg radio immunoassay (RIA) in the dilution test. HBcrAg concentrations correlated well with HBV-DNA TMA (r = 0.91, n = 29) and in-house real-time detection-PCR (r = 0.93, n = 47) in hepatitis B patients. On HBeAg/anti-HBe antibody seroconversion panels, the HBcrAg concentration changed in accordance with HBV-DNA levels. HBcrAg concentration provides a reflection of HBV virus load equivalent to HBV-DNA level, and the assay therefore offers a simple method for monitoring hepatitis B patients.
Figures


References
-
- Baker, B. L., A. M. D. Stecle, I. Peenze, and M. J. Mphahlele. 1995. Detection and quantitation of hepatitis B virus DNA: comparison of two commercial hybridization assays with polymerase chain reaction. J. Viral Hepatitis 2:107-111. - PubMed
-
- Bonino, F., F. Rosina, M. Rizzetto, R. Rizzi, E. Chiaberge, R. Tardanico, F. Callea, and G. Verme. 1986. Chronic hepatitis in HBsAg carriers with serum HBV-DNA and anti-HBe. Gastroenterology 90:1268-1273. - PubMed
-
- Carman, W. F., and H. C. Thomas. 1992. Genetic variation in hepatitis B virus. Gastroenterology 102:711-719. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

