Molecular diagnosis of clear cell sarcoma: detection of EWS-ATF1 and MITF-M transcripts and histopathological and ultrastructural analysis of 12 cases
- PMID: 11826187
- PMCID: PMC1906974
- DOI: 10.1016/S1525-1578(10)60679-4
Molecular diagnosis of clear cell sarcoma: detection of EWS-ATF1 and MITF-M transcripts and histopathological and ultrastructural analysis of 12 cases
Abstract
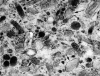
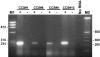
Clear cell sarcoma (CCS), also known as melanoma of soft parts, is an uncommon deep soft tissue tumor presenting typically in the lower extremities of young adults. Previous cytogenetic studies have established the specificity of the recurrent t(12;22)(q13;q12), resulting in a EWS-ATF1 fusion, for CCS. The prevalence of the EWS-ATF1 fusion in CCS remains unclear, since most genetically confirmed CCS have been reported as isolated cytogenetic or molecular diagnostic case reports. We therefore studied histologically confirmed CCS from 12 patients for the presence of EWS-ATF1 by reverse-transcriptase polymerase chain reaction (RT-PCR), using RNA extracted from either frozen (four cases) or formalin-fixed paraffin-embedded (eight cases) material. All primary tumors were located in the deep soft tissues of the extremities. Histologically, 10 cases had a typical epithelioid nested appearance. Most or all cases showed immunostaining for HMB45 (12 of 12), S-100 protein (10 of 12), and MITF (12 of 12). Ultrastructural analysis showed melanosomes in six of seven cases. The presence of an EWS-ATF1 fusion transcript was identified by RT-PCR in 11 of 12 cases (91%), all of which showed the same fusion transcript structure, namely the previously described in-frame fusion of EWS exon 8 to ATF1 codon 65. RT-PCR analysis for the melanocyte-specific splice form of the MITF transcript was positive in all cases tested (4 of 4). These data confirm that EWS-ATF1 detection can be used as a highly sensitive diagnostic test for CCS and that CCS expresses the melanocyte-specific form of the MITF transcript, further supporting its genuine melanocytic differentiation.
Figures





References
-
- Enzinger FM: Clear cell sarcoma of tendons and aponeuroses: an analysis of 21 cases. Cancer 1965, 18:1163-1174 - PubMed
-
- Chung EB, Enzinger FM: Malignant melanoma of soft parts: a reassessment of clear cell sarcoma. Am J Surg Pathol 1983, 7:403-413 - PubMed
-
- Kindblom L-G, Lodding O, Angervall L: Clear cell sarcoma of tendons and aponeuroses: an immunohistochemical and electron microscopic analysis indicating neural crest origin. Virchows Arch 1983, 401:109-128 - PubMed
-
- Benson JD, Kraemer B, Mackay B: Malignant melanoma of soft parts: an ultrastructural study of four cases. Ultrastruct Pathol 1985, 8:57-70 - PubMed
-
- Hasegawa T, Hirose T, Kudo E, Hizawa K: Clear cell sarcoma: an immunohistochemical and ultrastructural study. Acta Pathol Jpn 1989, 39:321-327 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

