SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis
- PMID: 11826297
- PMCID: PMC150549
- DOI: 10.1105/tpc.010215
SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis
Abstract
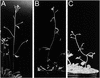
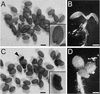
In higher plants, the shoot and the root generally show negative and positive gravitropism, respectively. To elucidate the molecular mechanisms involved in gravitropism, we have isolated many shoot gravitropism mutants in Arabidopsis. The sgr2 and zig/sgr4 mutants exhibited abnormal gravitropism in both inflorescence stems and hypocotyls. These genes probably are involved in the early step(s) of the gravitropic response. The sgr2 mutants also had misshapen seed and seedlings, whereas the stem of the zig/sgr4 mutants elongated in a zigzag fashion. The SGR2 gene encodes a novel protein that may be part of a gene family represented by bovine phosphatidic acid-preferring phospholipase A1 containing a putative transmembrane domain. This gene family has been reported only in eukaryotes. The ZIG gene was found to encode AtVTI11, a protein that is homologous with yeast VTI1 and is involved in vesicle transport. Our observations suggest that the two genes may be involved in a vacuolar membrane system that affects shoot gravitropism.
Figures

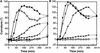




References
-
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Belyavskaya, N.A. (1996). Calcium and graviperception in plants: Inhibitor analysis. Int. Rev. Cytol. 168, 123–185.
-
- Bennett, M.J., Marchant, A., Green, H.G., May, S.T., Ward, S.P., Millner, P.A., Walker, A.R., Schulz, B., and Feldmann, K.A. (1996). Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 273, 948–950. - PubMed
-
- Chapman, K.D. (1998). Phospholipase activity during plant growth and development and in response to environmental stress. Trends Plant Sci. 3, 419–426.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

