The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei
- PMID: 11826299
- PMCID: PMC150551
- DOI: 10.1105/tpc.010319
The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei
Abstract
The slender rice1 mutant (slr1) shows a constitutive gibberellin (GA) response phenotype. To investigate the mode of action of SLR1, we generated transgenic rice expressing a fusion protein consisting of SLR1 and green fluorescent protein (SLR1-GFP) and analyzed the phenotype of the transformants and the subcellular localization of GFP in vivo. SLR1-GFP worked in nuclei to repress the GA signaling pathway; its overproduction caused a dwarf phenotype. Application of GA(3) to SLR1-GFP overproducers induced GA actions such as shoot elongation, downregulation of GA 20-oxidase expression, and upregulation of SLR1 expression linked with the disappearance of the nuclear SLR1-GFP protein. We also performed domain analyses of SLR1 using transgenic plants overproducing different kinds of truncated SLR1 proteins. The analyses revealed that the SLR1 protein can be divided into four parts: a GA signal perception domain located at the N terminus, a regulatory domain for its repression activity, a dimer formation domain essential for signal perception and repression activity, and a repression domain at the C terminus. We conclude that GA signal transduction is regulated by the appearance or disappearance of the nuclear SLR1 protein, which is controlled by the upstream GA signal.
Figures
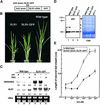
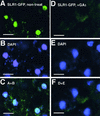
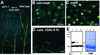

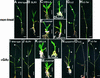
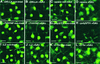
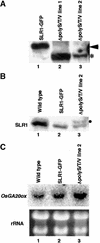

Comment in
-
Foolish seedlings and DELLA regulators: the functions of rice SLR1 and Arabidopsis RGL1 in GA signal transduction.Plant Cell. 2002 Jan;14(1):1-5. doi: 10.1105/tpc.140110. Plant Cell. 2002. PMID: 11826293 Free PMC article. No abstract available.
References
-
- Chakraborty, A., Saha, D., Bose, A., Chatterjee, M., and Gupta, N.K. (1994). Regulation of elF-2 α-subunit phosphorylation in reticulocyte lysate. Biochemistry 33, 6700–6706. - PubMed
-
- Chiu, W.-L., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. - PubMed
-
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 162, 156–159. - PubMed
-
- Chou, T.Y., Gerald, W.H., and Dang, C.V. (1995). c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J. Biol. Chem. 270, 18961–18965. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

