Arabidopsis RGL1 encodes a negative regulator of gibberellin responses
- PMID: 11826301
- PMCID: PMC150553
- DOI: 10.1105/tpc.010325
Arabidopsis RGL1 encodes a negative regulator of gibberellin responses
Abstract
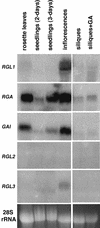
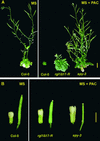
In Arabidopsis, the DELLA subfamily of GRAS regulatory genes consists of GAI, RGA, RGA-LIKE1 (RGL1), RGL2, and RGL3. GAI and RGA are known to be negative regulators of gibberellin (GA) responses. We found that RGL1 is a similar repressor of GA responses, as revealed by RGL1 gain-of-function and loss-of-function phenotypes. Repression of GA responses in Arabidopsis was conferred by a dominant 35S-rgl1 transgene carrying a DELLA domain deletion analogous to the GA-insensitive gai-1 mutation. As in GA-deficient Arabidopsis, the transgenic plants were dark green dwarfs with underdeveloped trichomes and flowers. Expression levels of GA4, a feedback-regulated GA biosynthetic gene, were increased correspondingly. Conversely, a loss-of-function rgl1 line had reduced GA4 expression and exhibited GA-independent activation of seed germination, leaf expansion, flowering, stem elongation, and floral development, as detected by resistance to the GA biosynthesis inhibitor paclobutrazol. RGL1 plays a greater role in seed germination than do GAI and RGA. The expression profile of RGL1 differed from those of the four other DELLA homologs. RGL1 message levels were predominant in flowers, with transcripts detected in developing ovules and anthers. As with RGA, green fluorescent protein (GFP)-tagged RGL1 protein was localized to the nucleus, but unlike GFP-RGA, there was no degradation after GA treatment. These findings indicate that RGL1 is a partially redundant, but distinct, negative regulator of GA responses and suggest that all DELLA subfamily members might possess separate as well as overlapping roles in GA signaling.
Figures






Comment in
-
Foolish seedlings and DELLA regulators: the functions of rice SLR1 and Arabidopsis RGL1 in GA signal transduction.Plant Cell. 2002 Jan;14(1):1-5. doi: 10.1105/tpc.140110. Plant Cell. 2002. PMID: 11826293 Free PMC article. No abstract available.
References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- Appel, R.D., Bairoch, A., and Hochstrasser, D.F. (1994). A new generation of information retrieval tools for biologists: The example of the ExPASy WWW server. Trends Biochem. Sci. 19, 258–260. - PubMed
-
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. - PubMed
-
- Bethke, P.C., and Jones, R.L. (1998). Gibberellin signaling. Curr. Opin. Plant Biol. 1, 440–446. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

