Arabidopsis CAP regulates the actin cytoskeleton necessary for plant cell elongation and division
- PMID: 11826305
- PMCID: PMC150557
- DOI: 10.1105/tpc.010301
Arabidopsis CAP regulates the actin cytoskeleton necessary for plant cell elongation and division
Abstract
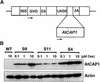
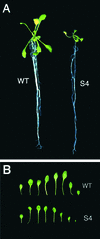
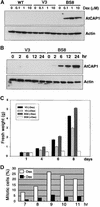
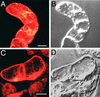
An Arabidopsis cDNA (AtCAP1) that encodes a predicted protein of 476 amino acids highly homologous with the yeast cyclase-associated protein (CAP) was isolated. Expression of AtCAP1 in the budding yeast CAP mutant was able to rescue defects such as abnormal cell morphology and random budding pattern. The C-terminal domain, 158 amino acids of AtCAP1 possessing in vitro actin binding activity, was needed for the regulation of cytoskeleton-related defects of yeast. Transgenic plants overexpressing AtCAP1 under the regulation of a glucocorticoid-inducible promoter showed different levels of AtCAP1 accumulation related to the extent of growth abnormalities, in particular size reduction of leaves as well as petioles. Morphological alterations in leaves were attributable to decreased cell size and cell number in both epidermal and mesophyll cells. Tobacco suspension-cultured cells (Bright Yellow 2) overexpressing AtCAP1 exhibited defects in actin filaments and were unable to undergo mitosis. Furthermore, an immunoprecipitation experiment suggested that AtCAP1 interacted with actin in vivo. Therefore, AtCAP1 may play a functional role in actin cytoskeleton networking that is essential for proper cell elongation and division.
Figures





References
-
- Aoyama, T., and Chua, N.H. (1997). A glucocorticoid-mediated transcription factor induction system in transgenic plants. Plant J. 11, 605–612. - PubMed
-
- Baluska, F., Jasik, J., Edelmann, H.G., Salajová, T., and Volkmann, D. (2001). Latriculin B-induced plant dwarfism: Plant cell elongation is F-actin-dependent. Dev. Biol. 231, 113–124. - PubMed
-
- Baum, B., Li, W., and Perrimon, N. (2000). A cyclase-associated protein regulates actin and cell polarity during Drosophila oogenesis and in yeast. Curr. Biol. 10, 964–973. - PubMed
-
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199.
-
- Benlali, A., Draskovic, I., Hazelett, D.J., and Treisman, J.E. (2000). act up controls actin polymerization to alter cell shape and restrict Hedgehog signaling in the Drosophila eye disc. Cell 101, 271–281. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

