Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction
- PMID: 11826309
- PMCID: PMC150561
- DOI: 10.1105/tpc.010304
Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction
Abstract
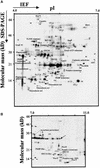
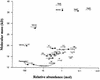
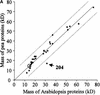
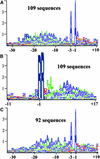
Experimental proteome analysis was combined with a genome-wide prediction screen to characterize the protein content of the thylakoid lumen of Arabidopsis chloroplasts. Soluble thylakoid proteins were separated by two-dimensional electrophoresis and identified by mass spectrometry. The identities of 81 proteins were established, and N termini were sequenced to validate localization prediction. Gene annotation of the identified proteins was corrected by experimental data, and an interesting case of alternative splicing was discovered. Expression of a surprising number of paralogs was detected. Expression of five isomerases of different classes suggests strong (un)folding activity in the thylakoid lumen. These isomerases possibly are connected to a network of peripheral and lumenal proteins involved in antioxidative response, including peroxiredoxins, m-type thioredoxins, and a lumenal ascorbate peroxidase. Characteristics of the experimentally identified lumenal proteins and their orthologs were used for a genome-wide prediction of the lumenal proteome. Lumenal proteins with a typical twin-arginine translocation motif were predicted with good accuracy and sensitivity and included additional isomerases and proteases. Thus, prime functions of the lumenal proteome include assistance in the folding and proteolysis of thylakoid proteins as well as protection against oxidative stress. Many of the predicted lumenal proteins must be present at concentrations at least 10,000-fold lower than proteins of the photosynthetic apparatus.
Figures









References
-
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Baier, M., Noctor, G., Foyer, C.H., and Dietz, K.J. (2000). Antisense suppression of 2-cysteine peroxiredoxin in Arabidopsis specifically enhances the activities and expression of enzymes associated with ascorbate metabolism but not glutathione metabolism. Plant Physiol. 124, 823–832. - PMC - PubMed
-
- Bassham, D.C., Creighton, A.M., Karnauchov, I., Herrmann, R.G., Klosgen, R.B., and Robinson, C. (1994). Mutations at the stromal processing peptidase cleavage site of a thylakoid lumen protein precursor affect the rate of processing but not the fidelity. J. Biol. Chem. 269, 16062–16066. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

