Repair of damaged DNA by Arabidopsis cell extract
- PMID: 11826311
- PMCID: PMC150563
- DOI: 10.1105/tpc.010258
Repair of damaged DNA by Arabidopsis cell extract
Abstract
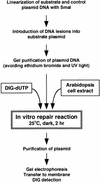
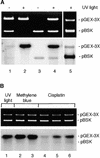
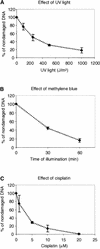
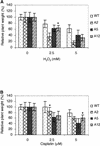
All living organisms have to protect the integrity of their genomes from a wide range of genotoxic stresses to which they are inevitably exposed. However, understanding of DNA repair in plants lags far behind such knowledge in bacteria, yeast, and mammals, partially as a result of the absence of efficient in vitro systems. Here, we report the experimental setup for an Arabidopsis in vitro repair synthesis assay. The repair of plasmid DNA treated with three different DNA-damaging agents, UV light, cisplatin, and methylene blue, after incubation with whole-cell extract was monitored. To validate the reliability of our assay, we analyzed the repair proficiency of plants depleted in AtRAD1 activity. The reduced repair of UV light- and cisplatin-damaged DNA confirmed the deficiency of these plants in nucleotide excision repair. Decreased repair of methylene blue-induced oxidative lesions, which are believed to be processed by the base excision repair machinery in mammalian cells, may indicate a possible involvement of AtRAD1 in the repair of oxidative damage. Differences in sensitivity to DNA polymerase inhibitors (aphidicolin and dideoxy TTP) between plant and human cell extracts were observed with this assay.
Figures


Similar articles
-
AtRAD1, a plant homologue of human and yeast nucleotide excision repair endonucleases, is involved in dark repair of UV damages and recombination.Plant J. 2000 Mar;21(6):507-18. doi: 10.1046/j.1365-313x.2000.00694.x. Plant J. 2000. PMID: 10758501
-
AtPolλ, a homolog of mammalian DNA polymerase λ in Arabidopsis thaliana, is involved in the repair of UV-B induced DNA damage through the dark repair pathway.Plant Cell Physiol. 2011 Feb;52(2):448-67. doi: 10.1093/pcp/pcr002. Epub 2011 Jan 11. Plant Cell Physiol. 2011. PMID: 21227935
-
Components of nucleotide excision repair and DNA damage tolerance in Arabidopsis thaliana.Environ Mol Mutagen. 2005 Mar-Apr;45(2-3):115-27. doi: 10.1002/em.20094. Environ Mol Mutagen. 2005. PMID: 15645454 Review.
-
Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity.EMBO J. 1999 Jun 15;18(12):3491-501. doi: 10.1093/emboj/18.12.3491. EMBO J. 1999. PMID: 10369688 Free PMC article.
-
Molecular mechanisms of DNA damage and repair: progress in plants.Crit Rev Biochem Mol Biol. 2001;36(4):337-97. doi: 10.1080/20014091074219. Crit Rev Biochem Mol Biol. 2001. PMID: 11563486 Review.
Cited by
-
Differential sensitivity of Arabidopsis siRNA biogenesis mutants to genotoxic stress.Plant Cell Rep. 2010 Dec;29(12):1401-10. doi: 10.1007/s00299-010-0930-9. Epub 2010 Oct 16. Plant Cell Rep. 2010. PMID: 20953786
-
Genome stability in the uvh6 mutant of Arabidopsis thaliana.Plant Cell Rep. 2014 Jun;33(6):979-91. doi: 10.1007/s00299-014-1580-0. Epub 2014 Feb 20. Plant Cell Rep. 2014. PMID: 24553752
-
ddm1 plants are sensitive to methyl methane sulfonate and NaCl stresses and are deficient in DNA repair.Plant Cell Rep. 2012 Sep;31(9):1549-61. doi: 10.1007/s00299-012-1269-1. Epub 2012 Apr 27. Plant Cell Rep. 2012. PMID: 22538524
-
CENTRIN2 interacts with the Arabidopsis homolog of the human XPC protein (AtRAD4) and contributes to efficient synthesis-dependent repair of bulky DNA lesions.Plant Mol Biol. 2006 May;61(1-2):345-56. doi: 10.1007/s11103-006-0016-9. Plant Mol Biol. 2006. PMID: 16786311
-
Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress.PLoS Genet. 2008 Jun 13;4(6):e1000093. doi: 10.1371/journal.pgen.1000093. PLoS Genet. 2008. PMID: 18551167 Free PMC article.
References
-
- Aboussekhra, A., Biggerstaff, M., Shivji, M.K.K., Vilpo, J.A., Moncollin, V., Podust, V.N., Protić, M., Hübscher, U., Egly, J.-M., and Wood, R.D. (1995). Mammalian DNA nucleotide excision repair reconstituted with purified components. Cell 80, 859–868. - PubMed
-
- Benedetto, J.P., Ech-Chaoui, R., Plissonneau, J., Laquel, P., Litvak, S., and Casroviejo, M. (1996). Changes of enzymes and factors involved in DNA synthesis during wheat germ germination. Plant Mol. Biol. 31, 1217–1225. - PubMed
-
- Bilang, R., Klöti, A., Schrot, M., and Potrykus, I. (1994). PEG-mediated direct gene transfer and electroporation. Plant Mol. Biol. Manual A1, 1–16.
-
- Britt, A.B. (1999). Molecular genetics of DNA repair in higher plants. Trends Plant Sci. 4, 20–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

