The yeast protein kinase Mps1p is required for assembly of the integral spindle pole body component Spc42p
- PMID: 11827982
- PMCID: PMC2173341
- DOI: 10.1083/jcb.200111025
The yeast protein kinase Mps1p is required for assembly of the integral spindle pole body component Spc42p
Abstract
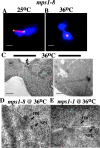
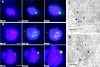
Saccharomyces cerevisiae MPS1 encodes an essential protein kinase that has roles in spindle pole body (SPB) duplication and the spindle checkpoint. Previously characterized MPS1 mutants fail in both functions, leading to aberrant DNA segregation with lethal consequences. Here, we report the identification of a unique conditional allele, mps1-8, that is defective in SPB duplication but not the spindle checkpoint. The mutations in mps1-8 are in the noncatalytic region of MPS1, and analysis of the mutant protein indicates that Mps1-8p has wild-type kinase activity in vitro. A screen for dosage suppressors of the mps1-8 conditional growth phenotype identified the gene encoding the integral SPB component SPC42. Additional analysis revealed that mps1-8 exhibits synthetic growth defects when combined with certain mutant alleles of SPC42. An epitope-tagged version of Mps1p (Mps1p-myc) localizes to SPBs and kinetochores by immunofluorescence microscopy and immuno-EM analysis. This is consistent with the physical interaction we detect between Mps1p and Spc42p by coimmunoprecipitation. Spc42p is a substrate for Mps1p phosphorylation in vitro, and Spc42p phosphorylation is dependent on Mps1p in vivo. Finally, Spc42p assembly is abnormal in a mps1-1 mutant strain. We conclude that Mps1p regulates assembly of the integral SPB component Spc42p during SPB duplication.
Figures







References
-
- Adams, I.R., and J.V. Kilmartin. 2000. Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 10:329–335. - PubMed
-
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smoth, and K. Struhl. 1997. Current protocols in molecular biology. John Wiley & Sons, Inc., New York. 50338 pp.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

