An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis
- PMID: 11830639
- PMCID: PMC122358
- DOI: 10.1073/pnas.032523999
An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis
Abstract
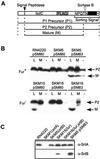
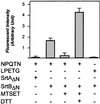
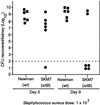
Sortase (SrtA), an enzyme that anchors surface proteins to the cell wall of Gram-positive bacteria, cleaves sorting signals at the LPXTG motif. We have identified a second sortase (SrtB) in the Gram-positive pathogen Staphylococcus aureus that is required for anchoring of a surface protein with a NPQTN motif. Purified SrtB cleaves NPQTN-bearing peptides in vitro, and a srtB mutant is defective in the persistence of animal infections. srtB is part of an iron-regulated locus called iron-responsive surface determinants (isd), which also contains a ferrichrome transporter and surface proteins with NPQTN and LPXTG motifs. Cell wall-anchored surface proteins and the isd locus seem involved in a novel mechanism of iron acquisition that is important for bacterial pathogenesis.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

