Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341
- PMID: 11830665
- PMCID: PMC122213
- DOI: 10.1073/pnas.032516399
Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341
Abstract
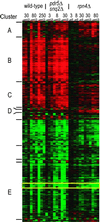
Although the biochemical targets of most drugs are known, the biological consequences of their actions are typically less well understood. In this study, we have used two whole-genome technologies in Saccharomyces cerevisiae to determine the cellular impact of the proteasome inhibitor PS-341. By combining population genomics, the screening of a comprehensive panel of bar-coded mutant strains, and transcript profiling, we have identified the genes and pathways most affected by proteasome inhibition. Many of these function in regulated protein degradation or a subset of mitotic activities. In addition, we identified Rpn4p as the transcription factor most responsible for the cell's ability to compensate for proteasome inhibition. Used together, these complementary technologies provide a general and powerful means to elucidate the cellular ramifications of drug treatment.
Figures



References
-
- Adams J, Palombella V J, Sausville E A, Johnson J, Destree A, Lazarus D D, Maas J, Pien C S, Prakash S, Elliott P J. Cancer Res. 1999;59:2615–2622. - PubMed
-
- Lee D, Goldberg A. J Biol Chem. 1996;271:27280–27284. - PubMed
-
- Lightcap E S, McCormack T A, Pien C S, Chau V, Adams J, Elliott P J. Clin Chem. 2000;46:673–683. - PubMed
-
- Elliott P J, McCormack T A, Pien C S, Adams J, Lightcap E S. Methods in Molecular Medicine. Totowa, NJ: Humana Press; 2002. , in press.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

