DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete
- PMID: 11830671
- PMCID: PMC122230
- DOI: 10.1073/pnas.032667699
DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete
Abstract
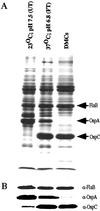
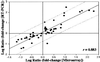
DNA microarrays were used to survey the adaptive genetic responses of Borrelia burgdorferi (Bb) B31, the Lyme disease spirochete, when grown under conditions analogous to those found in unfed ticks (UTs), fed ticks (FTs), or during mammalian host adaptation (Bb in dialysis membrane chambers implanted in rats). Microarrays contained 95.4% of the predicted B31 genes, 150 (8.6%) of which were differentially regulated (changes of > or = 1.8-fold) among the three growth conditions. A substantial proportion (46%) of the differentially regulated genes encoded proteins with predicted export signals (29% from predicted lipoproteins), emphasizing the importance to Bb of modulating its extracellular proteome. For B31 cultivated at the more restrictive UT condition, microarray data provided evidence of a bacterial stringent response and factors that restrict cell division. A large proportion of genes were responsive to the FT growth condition, wherein increased temperature and reduced pH were prominent environmental parameters. A surprising theme, supported by cluster analysis, was that many of the gene expression changes induced during the FT growth condition were transient and largely tempered as B31 adapted to the mammalian host, suggesting that once Bb gains entry and adapts to mammalian tissues, fewer differentially regulated genes are exploited. It therefore would seem that although widely dissimilar, the UT and dialysis membrane chamber growth conditions promote more static patterns of gene expression in Bb. The microarray data thus provide a basis for formulating new testable hypotheses regarding the life cycle of Bb and attaining a more complete understanding of many aspects of Bb's complex parasitic strategies.
Figures




References
-
- Steere A C. Hosp Pract. 1993;28:37–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

