From structure to function: YrbI from Haemophilus influenzae (HI1679) is a phosphatase
- PMID: 11835514
- PMCID: PMC3762886
- DOI: 10.1002/prot.10057
From structure to function: YrbI from Haemophilus influenzae (HI1679) is a phosphatase
Abstract
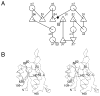
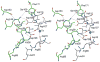
The crystal structure of the YrbI protein from Haemophilus influenzae (HI1679) was determined at a 1.67-A resolution. The function of the protein had not been assigned previously, and it is annotated as hypothetical in sequence databases. The protein exhibits the alpha/beta-hydrolase fold (also termed the Rossmann fold) and resembles most closely the fold of the L-2-haloacid dehalogenase (HAD) superfamily. Following this observation, a detailed sequence analysis revealed remote homology to two members of the HAD superfamily, the P-domain of Ca(2+) ATPase and phosphoserine phosphatase. The 19-kDa chains of HI1679 form a tetramer both in solution and in the crystalline form. The four monomers are arranged in a ring such that four beta-hairpin loops, each inserted after the first beta-strand of the core alpha/beta-fold, form an eight-stranded barrel at the center of the assembly. Four active sites are located at the subunit interfaces. Each active site is occupied by a cobalt ion, a metal used for crystallization. The cobalt is octahedrally coordinated to two aspartate side-chains, a backbone oxygen, and three solvent molecules, indicating that the physiological metal may be magnesium. HI1679 hydrolyzes a number of phosphates, including 6-phosphogluconate and phosphotyrosine, suggesting that it functions as a phosphatase in vivo. The physiological substrate is yet to be identified; however the location of the gene on the yrb operon suggests involvement in sugar metabolism.
Copyright 2002 Wiley‐Liss, Inc.
Figures





Similar articles
-
Structure of recombinant Haemophilus influenzae e (P4) acid phosphatase reveals a new member of the haloacid dehalogenase superfamily.Biochemistry. 2007 Oct 2;46(39):11110-9. doi: 10.1021/bi701016m. Epub 2007 Sep 8. Biochemistry. 2007. PMID: 17824671
-
Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 A resolution.Structure. 2001 Jan 10;9(1):65-71. doi: 10.1016/s0969-2126(00)00558-x. Structure. 2001. PMID: 11342136
-
YbiV from Escherichia coli K12 is a HAD phosphatase.Proteins. 2005 Mar 1;58(4):790-801. doi: 10.1002/prot.20267. Proteins. 2005. PMID: 15657928
-
An Atypical α/β-Hydrolase Fold Revealed in the Crystal Structure of Pimeloyl-Acyl Carrier Protein Methyl Esterase BioG from Haemophilus influenzae.Biochemistry. 2016 Dec 6;55(48):6705-6717. doi: 10.1021/acs.biochem.6b00818. Epub 2016 Nov 21. Biochemistry. 2016. PMID: 27933801
-
Human HAD phosphatases: structure, mechanism, and roles in health and disease.FEBS J. 2013 Jan;280(2):549-71. doi: 10.1111/j.1742-4658.2012.08633.x. Epub 2012 Jun 13. FEBS J. 2013. PMID: 22607316 Review.
Cited by
-
Neutron diffraction studies towards deciphering the protonation state of catalytic residues in the bacterial KDN9P phosphatase.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Sep;69(Pt 9):1015-9. doi: 10.1107/S1744309113021386. Epub 2013 Aug 19. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 23989152 Free PMC article.
-
Architecture and function of yeast phosphatidate phosphatase Pah1 domains/regions.Biochim Biophys Acta Mol Cell Biol Lipids. 2024 Dec;1869(8):159547. doi: 10.1016/j.bbalip.2024.159547. Epub 2024 Aug 3. Biochim Biophys Acta Mol Cell Biol Lipids. 2024. PMID: 39103045 Free PMC article. Review.
-
Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.Biochemistry. 2013 Aug 13;52(32):5372-86. doi: 10.1021/bi400659k. Epub 2013 Jul 29. Biochemistry. 2013. PMID: 23848398 Free PMC article.
-
A particle size threshold governs diffusion and segregation of PAR-3 during cell polarization.Cell Rep. 2022 Apr 12;39(2):110652. doi: 10.1016/j.celrep.2022.110652. Cell Rep. 2022. PMID: 35417695 Free PMC article.
-
The tail of KdsC: conformational changes control the activity of a haloacid dehalogenase superfamily phosphatase.J Biol Chem. 2009 Oct 30;284(44):30594-603. doi: 10.1074/jbc.M109.012278. Epub 2009 Sep 2. J Biol Chem. 2009. PMID: 19726684 Free PMC article.
References
-
- Selengut JD, Levine RL. MDP-1: A novel eukaryotic magnesium dependent phosphatase. Biochemistry. 2000;39:8315–8324. - PubMed
-
- Collet JF, van Schaftingen E, Stroobant V. A new family of phosphotransferases related to P-type ATPases. Trends Biochem Sci. 1998;23:284. - PubMed
-
- Collet JF, Stroobant V, Pirard M, Delpierre G, Van Schaftingen E. A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J Biol Chem. 1998;273:14107–14112. - PubMed
-
- Collet JF, Stroobant V, Van Schaftingen E. Mechanistic studies of phosphoserine phosphatase, an enzyme related to P-type AT-Pases. J Biol Chem. 1999;274:33985–33990. - PubMed
-
- Morais MC, Zhang W, Baker AS, Zhang G, Dunaway-Mariano D, Allen KN. The crystal structure of bacillus cereus phosphonoacetaldehyde hydrolase: insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily. Biochemistry. 2000;39:10385–10396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

