Human papillomavirus type 31 replication modes during the early phases of the viral life cycle depend on transcriptional and posttranscriptional regulation of E1 and E2 expression
- PMID: 11836404
- PMCID: PMC153800
- DOI: 10.1128/jvi.76.5.2263-2273.2002
Human papillomavirus type 31 replication modes during the early phases of the viral life cycle depend on transcriptional and posttranscriptional regulation of E1 and E2 expression
Abstract
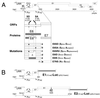
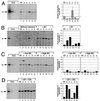
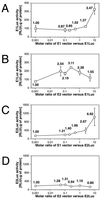
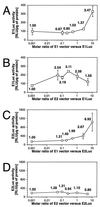
The E1 and E2 proteins are both required for papillomavirus DNA replication, and replication efficiency is controlled by the abundance of these factors. In human papillomaviruses (HPVs), the regulation of E1 and E2 expression and its effect on viral replication are not well understood. In particular, it is not known if E1 and E2 modulate their own expression and how posttranscriptional mechanisms may affect the levels of the replication proteins. Previous studies have implicated splicing within the E6 open reading frame (ORF) as being important for modulating replication of HPV type 31 (HPV31) through altered expression of E1 and E2. To analyze the function of the E6 intron in viral replication more specifically, we examined the effects of E6 splicing mutations in the context of entire viral genomes in transient assays. HPV31 genomes which had mutations in the splice donor site (E6SD) or the splice acceptor site (E6SA), a deletion of the intron (E6ID), or substituted heterologous intron sequences (E6IS) were constructed. Compared to wild-type (wt) HPV31, pHPV31-E6SD, -E6SA, and -E6IS replicated inefficiently while pHPV31-E6ID replicated at an intermediate level. Cotransfection of the E6 mutant genomes with an E1 expression vector strongly activated their replication levels, indicating that efficient expression of E1 requires E6 internal splicing. In contrast, replication was activated only moderately with an E2 expression vector. Replacing the wt E6 intron in HPV31 with a heterologous intron from simian virus 40 (E6SR2) resulted in replication levels similar to that of the wt in the absence of expression vectors, suggesting that mRNA splicing upstream of the E1 ORF is important for high-level replication. To examine the effects of E6 intron splicing on E1 and E2 expression directly, we constructed reporter DNAs in which the luciferase coding sequences were fused in frame to the E1 (E1Luc) or E2 (E2Luc) gene. Reporter activities were then analyzed in transient assays with cotransfected E1 or E2 expression vectors. Both reporters were moderately activated by E1 in a dose-dependent manner. In addition, E1Luc was activated by low doses of E2 but was repressed at high doses. In contrast, E2 had little effect on E2Luc activity. These data indicate that E1 expression and that of E2 are interdependent and regulated differentially. When the E6 splicing mutations were analyzed in both reporter backgrounds, only E1Luc activities correlated with splicing competence in the E6 ORF. These findings support the hypothesis that the E6 intron primarily regulates expression of E1. Finally, in long-term replication assays, none of the E6 mutant genomes could be stably maintained. However, cotransfection of the E6 splicing mutant genomes with pHPV31-E7NS, which contains a nonsense mutation in the E7 coding sequence, restored stable replication of some mutants. Our observations indicate that E1 expression and that of E2 are differentially regulated at multiple levels and that efficient expression of E1 is required for transient and stable viral replication. These regulatory mechanisms likely act to control HPV copy number during the various phases of the viral life cycle.
Figures

Similar articles
-
The E8E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes.J Virol. 2000 Feb;74(3):1178-86. doi: 10.1128/jvi.74.3.1178-1186.2000. J Virol. 2000. PMID: 10627528 Free PMC article.
-
Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes.Proc Natl Acad Sci U S A. 1999 Jul 20;96(15):8449-54. doi: 10.1073/pnas.96.15.8449. Proc Natl Acad Sci U S A. 1999. PMID: 10411895 Free PMC article.
-
A novel splice donor site at nt 1534 is required for long-term maintenance of HPV31 genomes.Virology. 2008 Jan 5;370(1):93-101. doi: 10.1016/j.virol.2007.08.026. Epub 2007 Sep 29. Virology. 2008. PMID: 17904182
-
Control of viral replication and transcription by the papillomavirus E8^E2 protein.Virus Res. 2017 Mar 2;231:96-102. doi: 10.1016/j.virusres.2016.11.005. Epub 2016 Nov 4. Virus Res. 2017. PMID: 27825778 Review.
-
Alternative splicing in the genome of HPV and its regulation.Front Cell Infect Microbiol. 2024 Oct 22;14:1443868. doi: 10.3389/fcimb.2024.1443868. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39502170 Free PMC article. Review.
Cited by
-
The binding of histone deacetylases and the integrity of zinc finger-like motifs of the E7 protein are essential for the life cycle of human papillomavirus type 31.J Virol. 2004 Apr;78(7):3533-41. doi: 10.1128/jvi.78.7.3533-3541.2004. J Virol. 2004. PMID: 15016876 Free PMC article.
-
HPV-18 E6 mutants reveal p53 modulation of viral DNA amplification in organotypic cultures.Proc Natl Acad Sci U S A. 2013 May 7;110(19):7542-9. doi: 10.1073/pnas.1304855110. Epub 2013 Apr 9. Proc Natl Acad Sci U S A. 2013. PMID: 23572574 Free PMC article.
-
Papillomavirus genome structure, expression, and post-transcriptional regulation.Front Biosci. 2006 Sep 1;11:2286-302. doi: 10.2741/1971. Front Biosci. 2006. PMID: 16720315 Free PMC article. Review.
-
Functional mapping of the human papillomavirus type 16 E1 cistron.J Virol. 2008 Nov;82(21):10724-34. doi: 10.1128/JVI.00921-08. Epub 2008 Aug 27. J Virol. 2008. PMID: 18753208 Free PMC article.
-
Role of the PDZ domain-binding motif of the oncoprotein E6 in the pathogenesis of human papillomavirus type 31.J Virol. 2004 Nov;78(22):12366-77. doi: 10.1128/JVI.78.22.12366-12377.2004. J Virol. 2004. PMID: 15507623 Free PMC article.
References
-
- Belaguli, N. S., M. M. Pater, and A. Pater. 1995. Splice sites of human papillomavirus type 16 E6 gene or heterologous gene required for transformation by E7 and accumulation of E7 RNA. J. Med. Virol. 47:445-453. - PubMed
-
- Chow, L. T., T. R. Broker, and J. B. Lewis. 1979. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J. Mol. Biol. 134:265-303. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

