The capsid of infectious bursal disease virus contains several small peptides arising from the maturation process of pVP2
- PMID: 11836417
- PMCID: PMC135936
- DOI: 10.1128/jvi.76.5.2393-2402.2002
The capsid of infectious bursal disease virus contains several small peptides arising from the maturation process of pVP2
Abstract
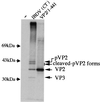
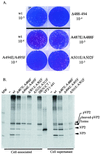
The capsid proteins VP2 and VP3 of infectious bursal disease virus, a birnavirus, are derived from the processing of a large polyprotein: NH2-pVP2-VP4-VP3-COOH. Although the primary cleavage sites at the pVP2-VP4 and VP4-VP3 junctions have been identified, the proteolytic cascade involved in the processing of this polyprotein is not yet fully understood, particularly the maturation of pVP2. By using different approaches, we showed that the processing of pVP2 (residues 1 to 512) generated VP2 and four small peptides (residues 442 to 487, 488 to 494, 495 to 501, and 502 to 512). We also showed that in addition to VP2, at least three of these peptides (residues 442 to 487, 488 to 494, and 502 to 512) were associated with the viral particles. The importance of the small peptides in the virus cycle was assessed by reverse genetics. Our results showed that the mutants lacking the two smaller peptides were viable, although the virus growth was affected. In contrast, deletions of the domain 442 to 487 or 502 to 512 did not allow virus recovery. Several amino acids of the peptide 502 to 512 appeared essential for virus viability. Substitutions of the P1 and/or P1" position were engineered at each of the cleavage sites (P1-P1": 441-442, 487-488, 494-495, 501-502, and 512-513). Most substitutions at the pVP2-VP4 junction (512-513) and at the final VP2 maturation cleavage site (441-442) were lethal. Mutations of intermediate cleavage sites (487-488, 494-495, and 501-502) led to viable viruses showing different but efficient pVP2 processing. Our data suggested that while peptides 488 to 494 and 495 to 501 play an accessory role, peptides 442 to 487 and 502 to 512 have an unknown but important function within the virus cycle.
Figures







References
-
- Azad, A. A., M. N. Jagadish, M. A. Brown, and P. J. Hudson. 1987. Deletion mapping and expression in Escherichia coli of the large genomic segment of a birnavirus. Virology 161:145-152. - PubMed
-
- Biacchesi, S., Y. X. Yu, M. Bearzotti, C. Tafalla, M. Fernandez-Alonso, and M. Bremont. 2000. Rescue of synthetic salmonid rhabdovirus minigenomes. J. Gen. Virol. 81:1941-1945. - PubMed
-
- Bong, D. T., C. Steinem, A. Janshoff, J. E. Johnson, and M. R. Ghadiri. 1999. A highly membrane-active peptide in Flock House virus: implications for the mechanism of nodavirus infection. Chem. Biol. 6:473-481. - PubMed
-
- Boot, H. J., A. A. H. M. ter Huurne, B. P. H. Peeters, and A. L. J. Gielkens. 1999. Efficient rescue of infectious bursal disease virus from cloned cDNA: evidence for involvement of the 3"-terminal sequence in genome replication. Virology 265:330-341. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

