The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds
- PMID: 11836534
- PMCID: PMC4035047
- DOI: 10.1038/nsb759
The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds
Abstract
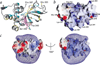
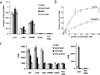
The PWWP domain is a weakly conserved sequence motif found in > 60 eukaryotic proteins, including the mammalian DNA methyltransferases Dnmt3a and Dnmt3b. These proteins often contain other chromatin-association domains. A 135-residue PWWP domain from mouse Dnmt3b (amino acids 223--357) has been structurally characterized at 1.8 A resolution. The N-terminal half of this domain resembles a barrel-like five-stranded structure, whereas the C-terminal half contains a five-helix bundle. The two halves are packed against each other to form a single structural module that exhibits a prominent positive electrostatic potential. The PWWP domain alone binds DNA in vitro, probably through its basic surface. We also show that recombinant Dnmt3b2 protein (a splice variant of Dnmt3b) and two N-terminal deletion mutants (Delta218 and Delta369) have approximately equal methyl transfer activity on unmethylated and hemimethylated CpG-containing oligonucleotides. The Delta218 protein, which includes the PWWP domain, binds DNA more strongly than Delta369, which lacks the PWWP domain.
Figures




References
-
- Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. - PubMed
-
- Wade PA. Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene. 2001;20:3166–3173. - PubMed
-
- Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. - PubMed
-
- Yoder JA, Soman NS, Verdine GL, Bestor TH. DNA (cytosine-5) methyltransferases in mouse cells and tisssues. Studies with a mechanism-based probe. J. Mol. Biol. 1997;270:385–395. - PubMed
-
- Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 1999;274:33002–33010. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
