Pathogenicity of missense and splice site mutations in hMSH2 and hMLH1 mismatch repair genes: implications for genetic testing
- PMID: 11839723
- PMCID: PMC1773142
- DOI: 10.1136/gut.50.3.405
Pathogenicity of missense and splice site mutations in hMSH2 and hMLH1 mismatch repair genes: implications for genetic testing
Abstract
Background: In hereditary non-polyposis colorectal cancer, over 90% of the identified mutations are in two genes, hMSH2 and hMLH1. A large proportion of the mutations detected in these genes are of the missense type which may be either deleterious mutations or harmless polymorphisms.
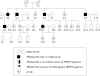
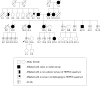
Aim: To investigate whether nine missense and one splice site mutation of hMLH1 and hMSH2, in 10 kindreds with a familial history of colorectal cancer or young age of onset, could be interpreted as pathogenic.
Methods: Clinical and genetic characteristics were collected: (i) evolutionary conservation of the codon involved; (ii) type of amino acid change; (iii) occurrence of mutation in healthy controls; (iv) cosegregation of mutation with disease phenotype; (v) functional consequences of gene variant; and (vi) microssatellite instability and immunoexpression of hMSH2 and hMLH1 analysis.
Results: Seven different missense and one splice site mutation were identified. Only 1/8 was found in the control group, 2/7 occurred in conserved residues, and 5/7 resulted in non-conservative changes. Functional studies were available for only 2/8 mutations. Segregation of the missense variant with disease phenotype was observed in three kindreds.
Conclusion: In the majority of families included, there was no definitive evidence that the missense or splice site alterations were causally associated with an increased risk of developing colorectal cancer. Until further evidence is available, these mutational events should be regarded and interpreted carefully and genetic diagnosis should not be offered to these kindreds.
Figures
References
-
- Marra G, Boland CR. Hereditary nonpolyposis colorectal cancer: the syndrome, the genes and historical perspectives. J Natl Cancer Inst 1995;87:1114–25. - PubMed
-
- Lynch HT, Smyrk T, Lynch JF. Overview of natural history, pathology, molecular genetics and management of HNPCC (Lynch syndrome). Int J Cancer 1996;69:38–43. - PubMed
-
- Vasen HFA, Wijnen JT, Menko FH, et al. Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology 1996;110:1020–7. - PubMed
-
- Watson P, Vasen HFA, Mecklin JP, et al. The risk of endometrial cancer in hereditary nonpolyposis colorectal cancer. Am J Med 1994;96:516–20. - PubMed
-
- Vasen HFA, Mecklin JP, Meera Khan P, et al. The International Collaborative Group on Hereditary non-polyposis Colorectal Cancer. Dis Colon Rectum 1991;34:424–5. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases