The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation
- PMID: 11842107
- PMCID: PMC100340
- DOI: 10.1093/nar/30.4.958
The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation
Abstract
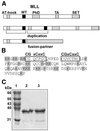
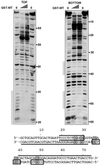
Alterations of the proto-oncogene MLL (mixed lineage leukemia) are characteristic for a high proportion of acute leukemias, especially those occurring in infants. The activation of MLL is achieved either by an internal tandem duplication of 5' MLL exons or by chromosomal translocations that create chimeric proteins with the N-terminus of MLL fused to a variety of different partner proteins. A domain of MLL with significant homology to the eukaryotic DNA methyltransferases (MT domain) has been found to be essential for the transforming potential of the oncogenic MLL derivatives. Here we demonstrate that this domain specifically recognizes DNA with unmethylated CpG sequences. In gel mobility shifts, the presence of CpG was sufficient for binding of recombinant GST-MT protein to DNA. The introduction of 5-methylCpG on one or both DNA strands precluded an efficient interaction. In surface plasmon resonance a KD of approximately 3.3 x 10(-8) M was determined for the GST-MT/DNA complex formation. Site selection experiments and DNase I footprinting confirmed CpG as the target of the MT domain. Finally, this interaction was corroborated in vivo in reporter assays utilizing the DNA-binding properties of the MT domain in a hybrid MT-VP16 transactivator construct.
Figures



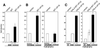
References
-
- Dimartino J.F. and Cleary,M.L. (1999) Mll rearrangements in haematological malignancies: lessons from clinical and biological studies. Br. J. Haematol., 106, 614–626. - PubMed
-
- Look A.T. (1997) Oncogenic transcription factors in the human acute leukemias. Science, 278, 1059–1064. - PubMed
-
- Tkachuk D.C., Kohler,S. and Cleary,M.L. (1992) Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell, 71, 691–700. - PubMed
-
- Pirrotta V. (1997) Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet., 13, 314–318. - PubMed
-
- Pirrotta V. (1998) Polycombing the genome: PcG, trxG and chromatin silencing. Cell, 93, 333–336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

