Complete disproportionation of duplex poly(dT)*poly(dA) into triplex poly(dT)*poly(dA)*poly(dT) and poly(dA) by coralyne
- PMID: 11842110
- PMCID: PMC100348
- DOI: 10.1093/nar/30.4.983
Complete disproportionation of duplex poly(dT)*poly(dA) into triplex poly(dT)*poly(dA)*poly(dT) and poly(dA) by coralyne
Abstract
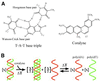
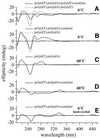
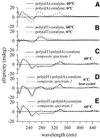
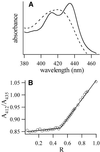
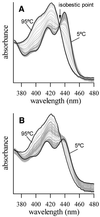
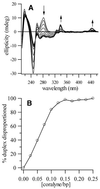
Coralyne is a small crescent-shaped molecule known to intercalate duplex and triplex DNA. We report that coralyne can cause the complete and irreversible disproportionation of duplex poly(dT)*poly(dA). That is, coralyne causes the strands of duplex poly(dT)*poly(dA) to repartition into equal molar equivalents of triplex poly(dT)*poly(dA)*poly(dT) and poly(dA). Poly(dT)*poly(dA) will remain as a duplex for months after the addition of coralyne, if the sample is maintained at 4 degrees C. However, disproportionation readily occurs upon heating above 35 degrees C and is not reversed by subsequent cooling. A titration of poly(dT)*poly(dA) with coralyne reveals that disproportionation is favored by as little as one molar equivalent of coralyne per eight base pairs of initial duplex. We have also found that poly(dA) forms a self-structure in the presence of coralyne with a melting temperature of 47 degrees C, for the conditions of our study. This poly(dA) self-structure binds coralyne with an affinity that is comparable with that of triplex poly(dT)*poly(dA)*poly(dT). A Job plot analysis reveals that the maximum level of poly(dA) self-structure intercalation is 0.25 coralyne molecules per adenine base. This conforms to the nearest neighbor exclusion principle for a poly(dA) duplex structure with A*A base pairs. We propose that duplex disproportionation by coralyne is promoted by both the triplex and the poly(dA) self-structure having binding constants for coralyne that are greater than that of duplex poly(dT)*poly(dA).
Figures


Similar articles
-
Controlling nucleic acid secondary structure by intercalation: effects of DNA strand length on coralyne-driven duplex disproportionation.Nucleic Acids Res. 2003 Aug 1;31(15):4608-15. doi: 10.1093/nar/gkg648. Nucleic Acids Res. 2003. PMID: 12888521 Free PMC article.
-
Coralyne binds tightly to both T.A.T- and C.G.C(+)-containing DNA triplexes.Biochemistry. 1993 Jun 1;32(21):5591-7. doi: 10.1021/bi00072a014. Biochemistry. 1993. PMID: 8504079
-
Investigation of DNA binding modes for a symmetrical cyanine dye trication: effect of DNA sequence and structure.J Biomol Struct Dyn. 2001 Jun;18(6):844-56. doi: 10.1080/07391102.2001.10506712. J Biomol Struct Dyn. 2001. PMID: 11444373
-
Poly(dA:dT) tracts: major determinants of nucleosome organization.Curr Opin Struct Biol. 2009 Feb;19(1):65-71. doi: 10.1016/j.sbi.2009.01.004. Epub 2009 Feb 7. Curr Opin Struct Biol. 2009. PMID: 19208466 Free PMC article. Review.
-
Isoquinoline alkaloids and their binding with polyadenylic acid: potential basis of therapeutic action.Mini Rev Med Chem. 2010 Jun;10(7):568-77. doi: 10.2174/138955710791384009. Mini Rev Med Chem. 2010. PMID: 20500148 Review.
Cited by
-
Hydration changes in the association of Hoechst 33258 with DNA.Biochemistry. 2005 Dec 27;44(51):16988-97. doi: 10.1021/bi051769x. Biochemistry. 2005. PMID: 16363812 Free PMC article.
-
Controlling nucleic acid secondary structure by intercalation: effects of DNA strand length on coralyne-driven duplex disproportionation.Nucleic Acids Res. 2003 Aug 1;31(15):4608-15. doi: 10.1093/nar/gkg648. Nucleic Acids Res. 2003. PMID: 12888521 Free PMC article.
-
Polymorphic nucleic Acid binding of bioactive isoquinoline alkaloids and their role in cancer.J Nucleic Acids. 2010;2010:593408. doi: 10.4061/2010/593408. Epub 2009 Dec 15. J Nucleic Acids. 2010. PMID: 20814427 Free PMC article.
-
A 'light-up' intercalator displacement assay for detection of triplex DNA stabilizers.Chem Commun (Camb). 2020 Feb 13;56(13):1996-1999. doi: 10.1039/c9cc08817b. Chem Commun (Camb). 2020. PMID: 31960843 Free PMC article.
-
Calorimetric and spectroscopic studies of aminoglycoside binding to AT-rich DNA triple helices.Biochimie. 2010 May;92(5):514-29. doi: 10.1016/j.biochi.2010.02.004. Epub 2010 Feb 16. Biochimie. 2010. PMID: 20167243 Free PMC article.
References
-
- Felsenfeld G. and Rich,A. (1957) Studies on the formation of two and three-stranded polyribonucleotides. Biochim. Biophys. Acta, 26, 457–468. - PubMed
-
- Mirkin S.M., Lyamichev,V.I., Drushlyak,K.N., Dobrynin,V.N., Filippov,S.A. and Frankkamenetskii,M.D. (1987) DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature, 330, 495–497. - PubMed
-
- Wells R.D., Collier,D.A., Hanvey,J.C., Shimizu,M. and Wohlrab,F. (1988) The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J., 2, 2939–2949. - PubMed
-
- Helene C. (1991) The anti-gene strategy: control of gene expression by triplex-forming-oligonucleotides. Anticancer Drug Des., 6, 569–584. - PubMed

