Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells
- PMID: 11842113
- PMCID: PMC100342
- DOI: 10.1093/nar/30.4.1009
Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells
Abstract
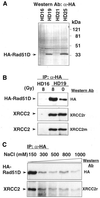
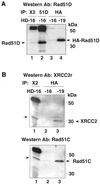
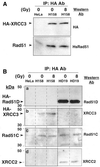
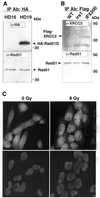
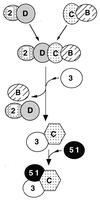
Genetic studies in rodent and chicken mutant cell lines have suggested that Rad51 paralogs (XRCC2, XRCC3, Rad51B/Rad51L1, Rad51C/Rad51L2 and Rad51D/Rad51L3) play important roles in homologous recombinational repair of DNA double-strand breaks and in maintaining chromosome stability. Previous studies using yeast two- and three-hybrid systems have shown interactions among these proteins, but it is not clear whether these interactions occur simultaneously or sequentially in vivo. By utilizing immunoprecipitation with extracts of human cells expressing epitope-tagged Rad51 paralogs, we demonstrate that XRCC2 and Rad51D, while stably interacting with each other, co-precipitate with Rad51C but not with XRCC3. In contrast, Rad51C is pulled down with XRCC3, whereas XRCC2 and Rad51D are not. In addition, Rad51B could be pulled down with Rad51C and Rad51D, but not with XRCC3. These results suggest that Rad51C is involved in two distinct in vivo complexes: Rad51B-Rad51C-Rad51D-XRCC2 and Rad51C-XRCC3. In addition, we demonstrate that Rad51 co-precipitates with XRCC3 but not with XRCC2 or Rad51D, suggesting that Rad51 can be present in an XRCC3-Rad51C-Rad51 complex. These complexes may act as functional units and serve accessory roles for Rad51 in the presynapsis stage of homologous recombinational repair.
Figures

References
-
- Shinohara A., Ogawa,H. and Ogawa,T. (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell, 69, 457–470. - PubMed
-
- Shinohara A., Ogawa,H., Matsuda,Y., Ushio,N., Ikeo,K. and Ogawa,T. (1993) Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nature Genet., 4, 239–243. - PubMed
-
- Sung P. (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science, 265, 1241–1243. - PubMed
-
- Baumann P., Benson,F.E. and West,S.C. (1996) Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell, 87, 757–766. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

