Neuroendocrine differentiation factor, IA-1, is a transcriptional repressor and contains a specific DNA-binding domain: identification of consensus IA-1 binding sequence
- PMID: 11842116
- PMCID: PMC100352
- DOI: 10.1093/nar/30.4.1038
Neuroendocrine differentiation factor, IA-1, is a transcriptional repressor and contains a specific DNA-binding domain: identification of consensus IA-1 binding sequence
Abstract
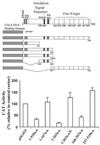
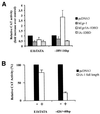
A novel cDNA, insulinoma-associated antigen-1 (IA-1), containing five zinc-finger DNA-binding motifs, was isolated from a human insulinoma subtraction library. IA-1 expression is restricted to fetal but not adult pancreatic and brain tissues as well as tumors of neuroendocrine origin. Using various GAL4 DNA binding domain (DBD)/IA-1 fusion protein constructs, we demonstrated that IA-1 functions as a transcriptional repressor and that the region between amino acids 168 and 263 contains the majority of the repressor activity. Using a selected and amplified random oligonucleotide binding assay and bacterially expressed GST-IA-1DBD fusion protein (257-510 a.a.), we identified the consensus IA-1 binding sequence, TG/TC/TC/TT/AGGGGG/TCG/A. Further experiments showed that zinc-fingers 2 and 3 of IA-1 are sufficient to demonstrate transcriptional activity using an IA-1 consensus site containing a reporter construct. A database search with the consensus IA-1 binding sequence revealed target sites in a number of pancreas- and brain-specific genes consistent with its restricted expression pattern. The most significant matches were for the 5'-flanking regions of IA-1 and NeuroD/beta2 genes. Co-transfection of cells with either the full-length IA-1 or hEgr-1AD/IA-1DBD construct and IA-1 or NeuroD/beta2 promoter/CAT construct modulated CAT activity. These findings suggest that the IA-1 protein may be auto-regulated and play a role in pancreas and neuronal development, specifically in the regulation of the NeuroD/beta2 gene.
Figures





References
-
- Goto Y., DeSilva,M.G., Toscani,A., Prabhakar,B.S., Notkins,A.L. and Lan,M.S. (1992) A novel human insulinoma-associated cDNA, IA-1, encodes a protein with zinc-finger DNA-binding motifs. J. Biol. Chem., 267, 15252–15257. - PubMed
-
- Lan M.S., Russell,E.K., Lu,J., Johnson,B.E. and Notkins,A.L. (1993) IA-1, a new marker for neuroendocrine differentiation in human lung cancer cell lines. Cancer Res., 53, 4169–4171. - PubMed
-
- Zhu M., Breslin,M.B. and Lan,M.S. (2001) A novel zinc-finger cDNA, IA-1, expression is associated with rat AR42J cell differentiation into insulin-positive cells. Pancreas, in press. - PubMed
-
- Huang H.P. and Tsai,M.J. (2000) Transcription factors involved in pancreatic islet development. J. Biomed. Sci., 7, 27–34. - PubMed
-
- Edlund H. (2001) Developmental biology of the pancreas. Diabetes, 50, S5–S9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

