Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6
- PMID: 11842152
- PMCID: PMC148911
- DOI: 10.1104/pp.010822
Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6
Abstract
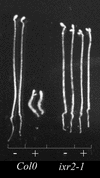
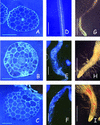
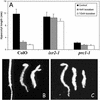
Isoxaben is a pre-emergence herbicide that inhibits cellulose biosynthesis in higher plants. Two loci identified by isoxaben-resistant mutants (ixr1-1, ixr1-2, and ixr2-1) in Arabidopsis have been reported previously. IXR1 was recently shown to encode the cellulose synthase catalytic subunit CESA3 (W.-R. Scheible, R. Eshed, T. Richmond, D. Delmer, and C. Somerville [2001] Proc Natl Acad Sci USA 98: 10079-10084). Here, we report on the cloning of IXR2, and show that it encodes another cellulose synthase isoform, CESA6. ixr2-1 carries a mutation substituting an amino acid close to the C terminus of CESA6 that is highly conserved among CESA family members. Transformation of wild-type plants with the mutated gene and not with the wild-type gene conferred increased resistance against the herbicide. The simplest interpretation for the existence of these two isoxaben-resistant loci is that CESA3 and CESA6 have redundant functions. However, loss of function procuste1 alleles of CESA6 were previously shown to have a strong growth defect and reduced cellulose content in roots and dark-grown hypocotyls. This indicates that in these mutants, the presence of CESA3 does not compensate for the absence of CESA6 in roots and dark-grown hypocotyls, which argues against redundant functions for CESA3 and CESA6. Together, these observations are compatible with a model in which CESA6 and CESA3 are active as a protein complex.
Figures


References
-
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. - PubMed
-
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thalianaplants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. - PubMed
-
- Bouchez D, Camilleri C. High molecular weight DNA extraction from Arabidopsis. Methods Mol Biol. 1998;82:61–70. - PubMed
-
- Delmer D. Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

