Engineering gene expression and protein synthesis by modulation of nuclear shape
- PMID: 11842191
- PMCID: PMC122304
- DOI: 10.1073/pnas.032668799
Engineering gene expression and protein synthesis by modulation of nuclear shape
Abstract
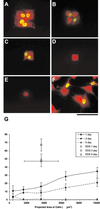
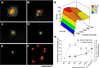
The current understanding of the relationships between cell shape, intracellular forces and signaling, nuclear shape and organization, and gene expression is in its infancy. Here we introduce a method for investigating gene-specific responses in individual cells with controlled nuclear shape and projected area. The shape of the nuclei of primary osteogenic cells were controlled on microfabricated substrata with regiospecific chemistry by confining attachment and spreading of isolated cells on adhesive islands. Gene expression and protein synthesis were altered by changing nuclear shape. Collagen I synthesis correlated directly with cell shape and nuclear shape index (NSI), where intermediate values of nuclear distension (6 < NSI < 8) promoted maximum synthesis. Osteocalcin mRNA, a bone-specific differentiation marker, was observed intracellularly by using reverse transcription in situ PCR at 4 days in cells constrained by the pattern and not detected in unconstrained cells of similar projected area, but different NSI. Our data supports the concept of gene expression and protein synthesis based on optimal distortion of the nucleus, possibly altering transcription factor affinity for DNA, transport to the nucleus, or nuclear matrix organization. The combination of microfabricated surfaces, reverse transcription in situ PCR, and NSI measurement is an excellent system to study how transcription factors, the nuclear matrix, and the cytoskeleton interact to control gene expression and may be useful for studying a wide variety of other cell shape/gene expression relationships.
Figures


References
-
- Chen C S, Mrksich M, Huang S, Whitesides G M, Ingber D E. Science. 1997;276:1425–1428. - PubMed
-
- Folkman J, Moscona A. Nature (London) 1978;273:345–349. - PubMed
-
- Singhvi R, Kumar A, Lopez G P, Stephanopoulos G N, Wang D I C, Whitesides G M, Ingbar D E. Science. 1994;264:696–698. - PubMed
-
- Aulthouse A L. Anat Rec. 1994;238:31–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

