Trinucleotide GAA repeats dictate pMGA gene expression in Mycoplasma gallisepticum by affecting spacing between flanking regions
- PMID: 11844762
- PMCID: PMC134842
- DOI: 10.1128/JB.184.5.1335-1339.2002
Trinucleotide GAA repeats dictate pMGA gene expression in Mycoplasma gallisepticum by affecting spacing between flanking regions
Abstract
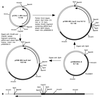
The pMGA genes of the avian respiratory pathogen Mycoplasma gallisepticum encode a family of hemagglutinins that are subject to phase variation. A trinucleotide GAA repeat region is located upstream of the pMGA transcription start site. The length of the repeat region varies at a high frequency due to changes in the number of repeat units. Previous studies have shown that pMGA genes are transcribed when 12 GAA repeats are present but are not transcribed when the number of repeats is not 12. To further analyze the mechanism of gene regulation, the pMGA promoter region was modified either by deleting the nucleotides 5" of the GAA repeats or by inserting linkers of 10 or 12 bp at a position 3" of the repeats. The modified promoter region was fused to a promoterless lacZ gene and transformed into M. gallisepticum by using transposon Tn4001 as a vector. Transformants and successive generations of progeny were analyzed with 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside (X-Gal) to monitor beta-galactosidase activity. For the transformants of M. gallisepticum containing the reporter with deletion of nucleotides 5" of the GAA repeats, GAA-dependent pMGA gene regulation was abolished. For the transformants containing the reporter with an addition of 10- or 12-bp linkers, lacZ was expressed only when eight GAA repeats were present. These data indicate that the nucleotides 5" of the GAA repeats as well as the spacing between the GAA repeats and sequences downstream (3") of the repeats are important for pMGA gene expression.
Figures




References
-
- Baseggio, N., M. D. Glew, P. F. Markham, K. G. Whithear, and G. F. Browning. 1996. Size and genomic location of the pMGA multigene family of Mycoplasma gallisepticum. Microbiology 142:1429-1435. - PubMed
-
- Dybvig, K., and L. L. Voelker. 1996. Molecular biology of mycoplasmas. Annu. Rev. Microbiol. 50:25-57. - PubMed
-
- Glew, M. D., P. F. Markham, G. F. Browning, and I. D. Walker. 1995. Expression studies on four members of the pMGA multigene family in Mycoplasma gallisepticum S6. Microbiology 141:3005-3014. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

