rRNA promoter activity in the fast-growing bacterium Vibrio natriegens
- PMID: 11844764
- PMCID: PMC134863
- DOI: 10.1128/JB.184.5.1349-1358.2002
rRNA promoter activity in the fast-growing bacterium Vibrio natriegens
Abstract
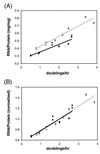
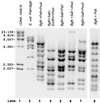
The bacterium Vibrio natriegens can double with a generation time of less than 10 min (R. G. Eagon, J. Bacteriol. 83:736-737, 1962), a growth rate that requires an extremely high rate of protein synthesis. We show here that V. natriegens' high potential for protein synthesis results from an increase in ribosome numbers with increasing growth rate, as has been found for other bacteria. We show that V. natriegens contains a large number of rRNA operons, and its rRNA promoters are extremely strong. The V. natriegens rRNA core promoters are at least as active in vitro as Escherichia coli rRNA core promoters with either E. coli RNA polymerase (RNAP) or V. natriegens RNAP, and they are activated by UP elements, as in E. coli. In addition, the E. coli transcription factor Fis activated V. natriegens rrn P1 promoters in vitro. We conclude that the high capacity for ribosome synthesis in V. natriegens results from a high capacity for rRNA transcription, and the high capacity for rRNA transcription results, at least in part, from the same factors that contribute most to high rates of rRNA transcription in E. coli, i.e., high gene dose and strong activation by UP elements and Fis.
Figures


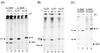

References
-
- Aiyar, S. E., S. M. McLeod, W. Ross, C. A. Hirvonen, M. S. Thomas, R. C. Johnson, and R. L. Gourse. 2002. Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J. Mol. Biol., in press. - PubMed
-
- Bag, P. K., S. Nandi, R. K. Bhadra, T. Ramamurthy, S. K. Bhattacharya, M. Nishibuchi, T. Hamabata, S. Yamasaki, Y. Takeda, and G. B. Nair. 1999. Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J. Clin. Microbiol. 37:2354-2357. - PMC - PubMed
-
- Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

