Positive growth rate-dependent regulation of the pdxA, ksgA, and pdxB genes of Escherichia coli K-12
- PMID: 11844765
- PMCID: PMC134838
- DOI: 10.1128/JB.184.5.1359-1369.2002
Positive growth rate-dependent regulation of the pdxA, ksgA, and pdxB genes of Escherichia coli K-12
Abstract
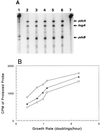
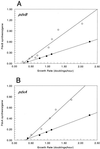
We found that transcription of the pdxA and pdxB genes, which mediate steps in the biosynthesis of the essential coenzyme pyridoxal 5"-phosphate, and the ksgA gene, which encodes an rRNA modification enzyme and is partly cotranscribed with pdxA, is subject to positive growth rate regulation in Escherichia coli K-12. The amounts of the pdxA-ksgA cotranscript and pdxB- and ksgA-specific transcripts and expression from pdxA- and pdxB-lacZ fusions increased as the growth rate increased. The half-lives of ksgA- and pdxB-specific transcripts were not affected by the growth rate, whereas the half-life of the pdxA-ksgA cotranscript was too short to be measured accurately. A method of normalization was applied to determine the amount of mRNA synthesized per gene and the rate of protein accumulation per gene. Normalization removed an apparent anomaly at fast growth rates and demonstrated that positive regulation of pdxB occurs at the level of transcription initiation over the whole range of growth rates tested. RNA polymerase limitation and autoregulation could not account for the positive growth rate regulation of pdxA, pdxB, and ksgA transcription. On the other hand, growth rate regulation of the amount of the pdxA-ksgA cotranscript was abolished by a fis mutation, suggesting a role for the Fis protein. In contrast, the fis mutation had no effect on pdxB- or ksgA-specific transcript amounts. The amounts of the pdxA-ksgA cotranscript and ksgA-specific transcript were repressed in the presence of high intracellular concentrations of guanosine tetraphosphate; however, this effect was independent of relA function for the pdxA-ksgA cotranscript. Amounts of the pdxB-specific transcript remained unchanged during amino acid starvation in wild-type and relA mutant strains.
Figures





References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smaith, and K. Struhl. 1993. Current protocols in molecular biology. Wiley-Interscience, New York, N.Y.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

