The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors
- PMID: 11847124
- PMCID: PMC125846
- DOI: 10.1093/emboj/21.4.769
The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors
Abstract
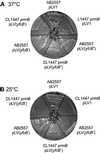
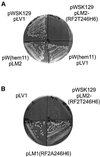
Class 1 peptide release factors (RFs) in Escherichia coli are N(5)-methylated on the glutamine residue of the universally conserved GGQ motif. One other protein alone has been shown to contain N(5)-methylglutamine: E.coli ribosomal protein L3. We identify the L3 methyltransferase as YfcB and show that it methylates ribosomes from a yfcB strain in vitro, but not RF1 or RF2. HemK, a close orthologue of YfcB, is shown to methylate RF1 and RF2 in vitro. hemK is immediately downstream of and co-expressed with prfA. Its deletion in E.coli K12 leads to very poor growth on rich media and abolishes methylation of RF1. The activity of unmethylated RF2 from K12 strains is extremely low due to the cumulative effects of threonine at position 246, in place of alanine or serine present in all other bacterial RFs, and the lack of N(5)-methylation of Gln252. Fast-growing spontaneous revertants in hemK K12 strains contain the mutations Thr246Ala or Thr246Ser in RF2. HemK and YfcB are the first identified methyltransferases modifying glutamine, and are widely distributed in nature.
Figures





Similar articles
-
HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination.Proc Natl Acad Sci U S A. 2002 Feb 5;99(3):1473-8. doi: 10.1073/pnas.032488499. Epub 2002 Jan 22. Proc Natl Acad Sci U S A. 2002. PMID: 11805295 Free PMC article.
-
A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation.EMBO J. 2000 Dec 15;19(24):6900-7. doi: 10.1093/emboj/19.24.6900. EMBO J. 2000. PMID: 11118225 Free PMC article.
-
The N5-glutamine S-adenosyl-L-methionine-dependent methyltransferase PrmC/HemK in Chlamydia trachomatis methylates class 1 release factors.J Bacteriol. 2005 Jan;187(2):507-11. doi: 10.1128/JB.187.2.507-511.2005. J Bacteriol. 2005. PMID: 15629922 Free PMC article.
-
[Identification and function of protein glutamine (N5) methyl transferase].Tanpakushitsu Kakusan Koso. 2003 May;48(6):733-9. Tanpakushitsu Kakusan Koso. 2003. PMID: 12816008 Review. Japanese. No abstract available.
-
The function, structure and regulation of E. coli peptide chain release factors.Biochimie. 1987 Oct;69(10):1031-41. doi: 10.1016/0300-9084(87)90003-4. Biochimie. 1987. PMID: 3126822 Review.
Cited by
-
NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs.Nature. 2015 Mar 19;519(7543):374-7. doi: 10.1038/nature14020. Epub 2014 Dec 22. Nature. 2015. PMID: 25533955
-
Low-molecular-weight protein tyrosine phosphatases of Bacillus subtilis.J Bacteriol. 2005 Jul;187(14):4945-56. doi: 10.1128/JB.187.14.4945-4956.2005. J Bacteriol. 2005. PMID: 15995210 Free PMC article.
-
COMBREX: a project to accelerate the functional annotation of prokaryotic genomes.Nucleic Acids Res. 2011 Jan;39(Database issue):D11-4. doi: 10.1093/nar/gkq1168. Epub 2010 Nov 21. Nucleic Acids Res. 2011. PMID: 21097892 Free PMC article.
-
Posttranslational protein modification in Archaea.Microbiol Mol Biol Rev. 2005 Sep;69(3):393-425. doi: 10.1128/MMBR.69.3.393-425.2005. Microbiol Mol Biol Rev. 2005. PMID: 16148304 Free PMC article. Review.
-
A Salmonella enterica serovar typhimurium hemA mutant is highly susceptible to oxidative DNA damage.J Bacteriol. 2002 Jul;184(14):3774-84. doi: 10.1128/JB.184.14.3774-3784.2002. J Bacteriol. 2002. PMID: 12081946 Free PMC article.
References
-
- Amann E., Ochs,B. and Abel,K.J. (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene, 69, 301–315. - PubMed
-
- Bailey T.L. and Elkan,C. (1994) Fitting a Mixture Model by Expectation Maximisation to Discover Motifs in Biopolymers. AAAI Press, Menlow Park, CA, pp. 28–36. - PubMed
-
- Blanc V. et al. (1997) Identification and analysis of genes from Streptomyces pristinaespiralis encoding enzymes involved in the biosynthesis of the 4-dimethylamino-l-phenylalanine precursor of pristinamycin I. Mol. Microbiol., 23, 191–202. - PubMed
-
- Bujnicki J.M. and Radlinska,M. (1999) Is the HemK family of putative S-adenosylmethionine-dependent methyltransferases a ‘missing’ ζ subfamily of adenine methyltransferases? A hypothesis. IUBMB Life, 48, 247–249. - PubMed
-
- Chang C.N. and Chang,F.N. (1975) Methylation of the ribosomal proteins in Escherichia coli. Nature and stoichiometry of the methylated amino acids in 50S ribosomal proteins. Biochemistry, 14, 468–477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

