The distinctive functions of the two structural calcium atoms in bovine pancreatic deoxyribonuclease
- PMID: 11847288
- PMCID: PMC2373464
- DOI: 10.1110/ps.20402
The distinctive functions of the two structural calcium atoms in bovine pancreatic deoxyribonuclease
Abstract
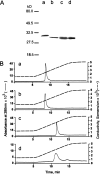
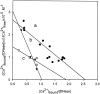
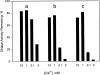
The two amino acid residues, Asp 99 and Asp 201, involved in the coordination of the two calcium atoms in the X-ray structure of bovine pancreatic (bp) DNase, were individually changed by site-directed mutagenesis. The two altered proteins, brDNase(D99A) and brDNase(D201A) were expressed in Escherichia coli and purified by anion exchange chromatography. Equilibrium dialysis showed that mutation destroyed one Ca(2+)-binding site each in brDNase(D99A) and brDNase(D201A). Compared with bpDNase, the Vmax value for brDNase(D99A) remained unchanged and that for brDNase(D201A) was decreased, whereas the K(m) values for the two variants were increased two- to threefold when the DNA hydrolytic hyperchromicity assay was used. Like bpDNase, brDNase(D99A) was able to make double scission on duplex DNA with Mg(2+) plus Ca(2+) and was effectively protected by Ca(2+) from the trypsin inactivation. But under the same conditions, brDNase(D201A) lost the double-scission ability and was not protected by Ca(2+). Nevertheless, the two variant proteins retained the characteristics of the Ca(2+)-induced conformational changes and the Ca(2+) protection against the beta-mercaptoethanol disruption of the essential disulfide bond, suggesting that other weaker Ca(2+)-binding sites not found in the X-ray structure were responsible for these properties. Therefore, the two structural calcium atoms are not for maintaining the overall conformation of the active DNase, as it has been indicated in the X-ray analysis, but rather play the role in the fine-tuning of the DNase activity.
Figures


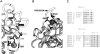

References
-
- Bradord, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. - PubMed
-
- Campbell, V.W. and Jackson, D.A. 1980. The effect of divalent cations on the mode of action of DNase I. J. Biol. Chem. 255 3726–3735. - PubMed
-
- Chen, C.Y., Lu, S.C., and Liao, T.H. 1998. Cloning, sequencing and expression in Escherichia coli of a DNA encoding bovine pancreatic deoxyribonuclease I: Purification and characterization of the recombinant enzyme. Gene 206181–184. - PubMed
-
- Corns, C.M. and Ludman, C.J. 1987. Some observations on the nature of the calcium-cresolphthalein complexon reaction and its relevance to the clinical laboratory. Ann. Clin. Bioche. 24 345–351. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

