Interference between D30N and L90M in selection and development of protease inhibitor-resistant human immunodeficiency virus type 1
- PMID: 11850252
- PMCID: PMC127489
- DOI: 10.1128/AAC.46.3.708-715.2002
Interference between D30N and L90M in selection and development of protease inhibitor-resistant human immunodeficiency virus type 1
Abstract
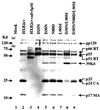
We studied the evolutionary relationships between the two protease inhibitor (PI) resistance mutations, D30N and L90M, of human immunodeficiency virus type 1 (HIV-1). The former is highly specific for nelfinavir resistance, while the latter is associated with resistance to several PIs, including nelfinavir. Among patients with nelfinavir treatment failure, we found that D30N acquisition was strongly suppressed when L90M preexisted. Thus, D30N/L90M double mutations not only were detected in a very limited number of patients but also accounted for a minor fraction within each patient. In the disease course, the D30N and L90M clones readily evolved independently of each other, and later the D30N/L90M double mutants emerged. The double mutants appeared to originate from the D30N lineage but not from the L90M lineage, or were strongly associated with the former. However, their evolutionary pathways appeared to be highly complex and to still have something in common, as they always contained several additional polymorphisms, including L63P and N88D, as common signatures. These results suggest that D30N and L90M are mutually exclusive during the evolutionary process. Supporting this notion, the D30N/L90M mutation was also quite rare in a large clinical database. Recombinant viruses with the relevant mutations were generated and compared for the ability to process p55gag and p160pol precursor proteins as well as for their infectivity. L90M caused little impairment of the cleavage activities, but D30N was detrimental, although significant residual activity was observed. In contrast, D30N/L90M demonstrated severe impairment. Thus, the concept of mutual antagonism of the two mutations was substantiated biochemically and functionally.
Figures

References
-
- Berkhout, B. 1999. HIV-1 evolution under pressure of protease inhibitors: climbing the stairs of viral fitness. J. Biomed. Sci. 6:298-305. - PubMed
-
- Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, D. Titus, T. Yang, H. Teppler, K. E. Squires, P. J. Deutsch, and E. A. Emini. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. - PubMed
-
- Debouck, C. 1992. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res. Hum. Retroviruses 8:153-164. - PubMed
-
- Gulnik, S. V., L. I. Suvorov, B. Liu, B. Yu, B. Anderson, H. Mitsuya, and J. W. Erickson. 1995. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry 34:9282-9287. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

