Oncogene cooperativity in Friend erythroleukemia: erythropoietin receptor activation by the env gene of SFFV leads to transcriptional upregulation of PU.1, independent of SFFV proviral insertion
- PMID: 11850847
- PMCID: PMC2388250
- DOI: 10.1038/sj.onc.1205183
Oncogene cooperativity in Friend erythroleukemia: erythropoietin receptor activation by the env gene of SFFV leads to transcriptional upregulation of PU.1, independent of SFFV proviral insertion
Abstract
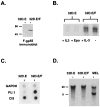
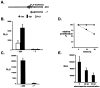
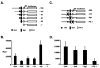
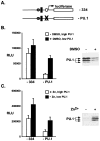
Cancer is a multi-step, multi-genetic event. Whether oncogenic mutations cooperate with one another to transform cells and how is not well understood. The Friend murine retroviral erythroleukemia model involves mitogenic activation of the erythropoietin receptor (EpoR) by the virus env gene (F-gp55), aberrant over-expression of the transcription factor PU.1, and inactivating mutations in p53. In this report we demonstrate that concurrent expression of F-gp55 and PU.1 in erythroid target cells, in vivo, cooperate to accelerate erythroleukemia induction. Early in the disease, prior to the detection of clonal leukemic cells, activation of the EpoR by F-gp55, but not erythropoietin, resulted in transcriptional upregulation of PU.1 through a trans regulatory mechanism. This could occur in the absence of an integrated provirus within the PU.1 gene locus. The regulation of PU.1 transcription in established erythroleukemia cell lines differed depending upon the level of PU.1 protein present. Our results suggest that the action of F-gp55 contributes to both early and late stages of Friend erythroleukemia and that persistence of F-gp55 expression may be required not only to initiate erythroleukemia but to also maintain erythroleukemia following Friend virus infection.
Figures


References
-
- Barnache S, Wendling F, Lacombe C, Denis N, Titeux M, Vainchecker W, Moreaeu-Gachelin F. Oncogene. 1998;16:2989–2995. - PubMed
-
- Beckman DL, Lin LL, Quinones ME, Longmore GD. Blood. 1999;94:2667–2675. - PubMed
-
- Ben-David Y, Bernstein A. Cell. 1991;66:831–834. - PubMed
-
- Chen H, Ray-Gallet D, Zhang P, Hetherington CJ, Gonzalez DA, Zhang D-E, Moreau-Gachelin F, Tenen DG. Oncogene. 1995;11:1549–1560. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

