Quorum-sensing regulators control virulence gene expression in Vibrio cholerae
- PMID: 11854465
- PMCID: PMC122484
- DOI: 10.1073/pnas.052694299
Quorum-sensing regulators control virulence gene expression in Vibrio cholerae
Abstract
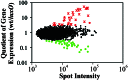
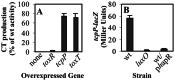
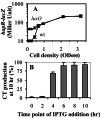
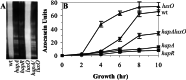
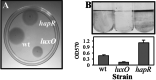
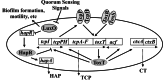
The production of virulence factors including cholera toxin and the toxin-coregulated pilus in the human pathogen Vibrio cholerae is strongly influenced by environmental conditions. The well-characterized ToxR signal transduction cascade is responsible for sensing and integrating the environmental information and controlling the virulence regulon. We show here that, in addition to the known components of the ToxR signaling circuit, quorum-sensing regulators are involved in regulation of V. cholerae virulence. We focused on the regulators LuxO and HapR because homologues of these two proteins control quorum sensing in the closely related luminous marine bacterium Vibrio harveyi. Using an infant mouse model, we found that a luxO mutant is severely defective in colonization of the small intestine. Gene arrays were used to profile transcription in the V. cholerae wild type and the luxO mutant. These studies revealed that the ToxR regulon is repressed in the luxO mutant, and that this effect is mediated by another negative regulator, HapR. We show that LuxO represses hapR expression early in log-phase growth, and constitutive expression of hapR blocks ToxR-regulon expression. Additionally, LuxO and HapR regulate a variety of other cellular processes including motility, protease production, and biofilm formation. Together these data suggest a role for quorum sensing in modulating expression of blocks of virulence genes in a reciprocal fashion in vivo.
Figures

Similar articles
-
Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter.Mol Microbiol. 2002 Nov;46(4):1135-47. doi: 10.1046/j.1365-2958.2002.03229.x. Mol Microbiol. 2002. PMID: 12421317
-
Vibrio cholerae CsrA Regulates ToxR Levels in Response to Amino Acids and Is Essential for Virulence.mBio. 2015 Aug 4;6(4):e01064. doi: 10.1128/mBio.01064-15. mBio. 2015. PMID: 26242626 Free PMC article.
-
The transcriptional regulator VqmA increases expression of the quorum-sensing activator HapR in Vibrio cholerae.J Bacteriol. 2006 Apr;188(7):2446-53. doi: 10.1128/JB.188.7.2446-2453.2006. J Bacteriol. 2006. PMID: 16547031 Free PMC article.
-
Regulation of virulence in Vibrio cholerae: the ToxR regulon.Future Microbiol. 2007 Jun;2(3):335-44. doi: 10.2217/17460913.2.3.335. Future Microbiol. 2007. PMID: 17661707 Review.
-
Quorum Sensing Gene Regulation by LuxR/HapR Master Regulators in Vibrios.J Bacteriol. 2017 Sep 5;199(19):e00105-17. doi: 10.1128/JB.00105-17. Print 2017 Oct 1. J Bacteriol. 2017. PMID: 28484045 Free PMC article. Review.
Cited by
-
The Cpx system regulates virulence gene expression in Vibrio cholerae.Infect Immun. 2015 Jun;83(6):2396-408. doi: 10.1128/IAI.03056-14. Epub 2015 Mar 30. Infect Immun. 2015. PMID: 25824837 Free PMC article.
-
Vibrio cholerae cytolysin recognizes the heptasaccharide core of complex N-glycans with nanomolar affinity.J Mol Biol. 2013 Mar 11;425(5):944-57. doi: 10.1016/j.jmb.2012.12.016. Epub 2012 Dec 28. J Mol Biol. 2013. PMID: 23274141 Free PMC article.
-
Toxin synthesis by Clostridium difficile is regulated through quorum signaling.mBio. 2015 Feb 24;6(2):e02569. doi: 10.1128/mBio.02569-14. mBio. 2015. PMID: 25714717 Free PMC article.
-
Bundle-forming pili and EspA are involved in biofilm formation by enteropathogenic Escherichia coli.J Bacteriol. 2006 Jun;188(11):3952-61. doi: 10.1128/JB.00177-06. J Bacteriol. 2006. PMID: 16707687 Free PMC article.
-
The intragenus and interspecies quorum-sensing autoinducers exert distinct control over Vibrio cholerae biofilm formation and dispersal.PLoS Biol. 2019 Nov 11;17(11):e3000429. doi: 10.1371/journal.pbio.3000429. eCollection 2019 Nov. PLoS Biol. 2019. PMID: 31710602 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

