A method for optimizing potential-energy functions by a hierarchical design of the potential-energy landscape: application to the UNRES force field
- PMID: 11854494
- PMCID: PMC122298
- DOI: 10.1073/pnas.032675399
A method for optimizing potential-energy functions by a hierarchical design of the potential-energy landscape: application to the UNRES force field
Abstract
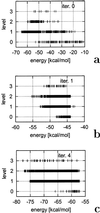
A method for optimizing potential-energy functions of proteins is proposed. The method assumes a hierarchical structure of the energy landscape, which means that the energy decreases as the number of native-like elements in a structure increases, being lowest for structures from the native family and highest for structures with no native-like element. A level of the hierarchy is defined as a family of structures with the same number of native-like elements (or degree of native likeness). Optimization of a potential-energy function is aimed at achieving such a hierarchical structure of the energy landscape by forcing appropriate free-energy gaps between hierarchy levels to place their energies in ascending order. This procedure is different from methods developed thus far, in which the energy gap and/or the Z score between the native structure and all non-native structures are maximized, regardless of the degree of native likeness of the non-native structures. The advantage of this approach lies in reducing the number of structures with decreasing energy, which should ensure the searchability of the potential. The method was tested on two proteins, PDB ID codes and, with an off-lattice united-residue force field. For, the search of the conformational space with the use of the conformational space annealing method and the newly optimized potential-energy function found the native structure very quickly, as opposed to the potential-energy functions obtained by former optimization methods. After even incomplete optimization, the force field obtained by using located the native-like structures of two peptides, and betanova (a designed three-stranded beta-sheet peptide), as the lowest-energy conformations, whereas for the 46-residue N-terminal fragment of staphylococcal protein A, the native-like conformation was the second-lowest-energy conformation and had an energy 2 kcal/mol above that of the lowest-energy structure.
Figures


Similar articles
-
Modification and optimization of the united-residue (UNRES) potential energy function for canonical simulations. I. Temperature dependence of the effective energy function and tests of the optimization method with single training proteins.J Phys Chem B. 2007 Jan 11;111(1):260-85. doi: 10.1021/jp065380a. J Phys Chem B. 2007. PMID: 17201450 Free PMC article.
-
Energy-based de novo protein folding by conformational space annealing and an off-lattice united-residue force field: application to the 10-55 fragment of staphylococcal protein A and to apo calbindin D9K.Proc Natl Acad Sci U S A. 1999 Mar 2;96(5):2025-30. doi: 10.1073/pnas.96.5.2025. Proc Natl Acad Sci U S A. 1999. PMID: 10051588 Free PMC article.
-
Ab initio simulations of protein-folding pathways by molecular dynamics with the united-residue model of polypeptide chains.Proc Natl Acad Sci U S A. 2005 Feb 15;102(7):2362-7. doi: 10.1073/pnas.0408885102. Epub 2005 Jan 26. Proc Natl Acad Sci U S A. 2005. PMID: 15677316 Free PMC article.
-
Design of a knowledge-based force field for off-lattice simulations of protein structure.Acta Biochim Pol. 1997;44(3):527-47. Acta Biochim Pol. 1997. PMID: 9511963 Review.
-
Coarse-grained force field: general folding theory.Phys Chem Chem Phys. 2011 Oct 14;13(38):16890-901. doi: 10.1039/c1cp20752k. Epub 2011 Jun 3. Phys Chem Chem Phys. 2011. PMID: 21643583 Free PMC article. Review.
Cited by
-
In silico Design of Laccase Thermostable Mutants From Lacc 6 of Pleurotus Ostreatus.Front Microbiol. 2018 Nov 14;9:2743. doi: 10.3389/fmicb.2018.02743. eCollection 2018. Front Microbiol. 2018. PMID: 30487785 Free PMC article.
-
Statistical mechanics-based method to extract atomic distance-dependent potentials from protein structures.Proteins. 2011 Sep;79(9):2648-61. doi: 10.1002/prot.23086. Epub 2011 Jul 5. Proteins. 2011. PMID: 21732421 Free PMC article.
-
Determination of side-chain-rotamer and side-chain and backbone virtual-bond-stretching potentials of mean force from AM1 energy surfaces of terminally-blocked amino-acid residues, for coarse-grained simulations of protein structure and folding. II. Results, comparison with statistical potentials, and implementation in the UNRES force field.J Comput Chem. 2010 Apr 30;31(6):1154-67. doi: 10.1002/jcc.21402. J Comput Chem. 2010. PMID: 20017135 Free PMC article.
-
Physics-based protein-structure prediction using a hierarchical protocol based on the UNRES force field: assessment in two blind tests.Proc Natl Acad Sci U S A. 2005 May 24;102(21):7547-52. doi: 10.1073/pnas.0502655102. Epub 2005 May 13. Proc Natl Acad Sci U S A. 2005. PMID: 15894609 Free PMC article.
-
Replica Exchange and Multicanonical Algorithms with the coarse-grained UNRES force field.J Chem Theory Comput. 2006;2(3):513-528. doi: 10.1021/ct050253o. J Chem Theory Comput. 2006. PMID: 18797518 Free PMC article.
References
-
- Vásquez M, Némethy G, Scheraga H A. Chem Rev. 1994;94:2183–2239.
-
- Anfinsen C B. Science. 1973;181:223–230. - PubMed
-
- Crippen G M, Snow E. Biopolymers. 1990;29:1479–1489. - PubMed
-
- Seetharamulu P, Crippen G M. J Math Chem. 1991;6:91–110.
-
- Sali A, Shakhnovich E I, Karplus M. Nature (London) 1994;369:248–251. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

